Transfer RNA supplementation rescues HARS deficiency in a humanized yeast model of Charcot-Marie-Tooth disease
- PMID: 39530218
- PMCID: PMC11662652
- DOI: 10.1093/nar/gkae996
Transfer RNA supplementation rescues HARS deficiency in a humanized yeast model of Charcot-Marie-Tooth disease
Abstract
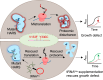
Aminoacyl-tRNA synthetases are indispensable enzymes in all cells, ensuring the correct pairing of amino acids to their cognate tRNAs to maintain translation fidelity. Autosomal dominant mutations V133F and Y330C in histidyl-tRNA synthetase (HARS) cause the genetic disorder Charcot-Marie-Tooth type 2W (CMT2W). Treatments are currently restricted to symptom relief, with no therapeutic available that targets the cause of disease. We previously found that histidine supplementation alleviated phenotypic defects in a humanized yeast model of CMT2W caused by HARS V155G and S356N that also unexpectedly exacerbated the phenotype of the two HARS mutants V133F and Y330C. Here, we show that V133F destabilizes recombinant HARS protein, which is rescued in the presence of tRNAHis. HARS V133F and Y330C cause mistranslation and cause changes to the proteome without activating the integrated stress response as validated by mass spectrometry and growth defects that persist with histidine supplementation. The growth defects and reduced translation fidelity caused by V133F and Y330C mutants were rescued by supplementation with human tRNAHis in a humanized yeast model. Our results demonstrate the feasibility of cognate tRNA as a therapeutic that rescues HARS deficiency and ameliorates toxic mistranslation generated by causative alleles for CMT.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Similar articles
-
Histidine supplementation can escalate or rescue HARS deficiency in a Charcot-Marie-Tooth disease model.Hum Mol Genet. 2023 Feb 19;32(5):810-824. doi: 10.1093/hmg/ddac239. Hum Mol Genet. 2023. PMID: 36164730 Free PMC article.
-
Towards a Cure for HARS Disease.Genes (Basel). 2023 Jan 18;14(2):254. doi: 10.3390/genes14020254. Genes (Basel). 2023. PMID: 36833180 Free PMC article. Review.
-
Characterization of a novel heterozygous variant in the histidyl-tRNA synthetase gene associated with Charcot-Marie-Tooth disease type 2W.IUBMB Life. 2024 Dec;76(12):1125-1138. doi: 10.1002/iub.2918. Epub 2024 Oct 1. IUBMB Life. 2024. PMID: 39352000 Free PMC article.
-
Substrate interaction defects in histidyl-tRNA synthetase linked to dominant axonal peripheral neuropathy.Hum Mutat. 2018 Mar;39(3):415-432. doi: 10.1002/humu.23380. Epub 2017 Dec 26. Hum Mutat. 2018. PMID: 29235198 Free PMC article.
-
Neurodegenerative Charcot-Marie-Tooth disease as a case study to decipher novel functions of aminoacyl-tRNA synthetases.J Biol Chem. 2019 Apr 5;294(14):5321-5339. doi: 10.1074/jbc.REV118.002955. Epub 2019 Jan 14. J Biol Chem. 2019. PMID: 30643024 Free PMC article. Review.
Cited by
-
Transfer RNA and small molecule therapeutics for aminoacyl-tRNA synthetase diseases.FEBS J. 2025 Jun;292(11):2737-2750. doi: 10.1111/febs.17361. Epub 2024 Dec 19. FEBS J. 2025. PMID: 39702998 Free PMC article. Review.
-
Yeast models for Charcot-Marie-Tooth disease-causing aminoacyl-tRNA synthetase alleles reveal the cellular basis of disease.IUBMB Life. 2025 Apr;77(4):e70017. doi: 10.1002/iub.70017. IUBMB Life. 2025. PMID: 40156251 Free PMC article. Review.
References
-
- Antonellis A., Ellsworth R.E., Sambuughin N., Puls I., Abel A., Lee-Lin S.Q., Jordanova A., Kremensky I., Christodoulou K., Middleton L.T.et al. .. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am. J. Hum. Genet. 2003; 72:1293–1299. - PMC - PubMed
-
- Jordanova A., Irobi J., Thomas F.P., Van Dijck P., Meerschaert K., Dewil M., Dierick I., Jacobs A., De Vriendt E., Guergueltcheva V.et al. .. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat. Genet. 2006; 38:197–202. - PubMed
-
- Latour P., Thauvin-Robinet C., Baudelet-Mery C., Soichot P., Cusin V., Faivre L., Locatelli M.C., Mayencon M., Sarcey A., Broussolle E.et al. .. A major determinant for binding and aminoacylation of tRNA (Ala) in cytoplasmic Alanyl-tRNA synthetase is mutated in dominant axonal Charcot-Marie-Tooth disease. Am. J. Hum. Genet. 2010; 86:77–82. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

