SRY+ Derivative X Chromosome in a Female With Apparently Typical Sexual Development
- PMID: 39530251
- PMCID: PMC11555487
- DOI: 10.1002/mgg3.70033
SRY+ Derivative X Chromosome in a Female With Apparently Typical Sexual Development
Abstract
Background: When the SRY gene is present in a 46,XX fetus, some degree of testicular development is expected. Our laboratory performed prenatal genetic testing for a fetus that had screened positive for Y chromosome material by noninvasive prenatal screening (NIPS) but that had apparently typical female development by ultrasound imaging. The aim of this study was to determine the clinical relevance of the NIPS results.
Methods: We analyzed fetal material obtained via amniocentesis procedure by G-banding, microarray, and fluorescence in situ hybridization (FISH). Optical genome mapping (OGM) was also performed.
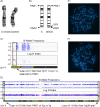
Results: G-band analysis revealed a normal 46,XX karyotype. Microarray and FISH analyses together detected an SRY+ gain of 5.7 Mb from terminal Yp that was translocated to terminal Xq, with a loss of 1.6 Mb from terminal Xq. The final karyotype was 46,X,der(X)t(X;Y)(q28;p11.2). Prenatal ultrasound and postnatal physical examination revealed apparently typical female genitalia. The Xq deletion encompassed a gene, IKBKG, that is sensitive to loss of function, suggesting that preferential inactivation of the derivative X chromosome allowed for typical female development. OGM software did not directly identify this translocation.
Conclusion: This case demonstrates how the SRY gene may be present in a 46,XX biological female without differences of sexual development.
Keywords: X inactivation; differences of sexual development; ovotesticular; sex reversal.
© 2024 The Author(s). Molecular Genetics & Genomic Medicine published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Non-Invasive Prenatal Test Analysis Opens a Pandora's Box: Identification of Very Rare Cases of SRY-Positive Healthy Females, Segregating for Three Generations Thanks to Preferential Inactivation of the XqYp Translocated Chromosome.Genes (Basel). 2024 Jan 16;15(1):103. doi: 10.3390/genes15010103. Genes (Basel). 2024. PMID: 38254992 Free PMC article.
-
Prenatal diagnosis of sex chromosomal inversion, translocation and deletion.Mol Med Rep. 2018 Feb;17(2):2811-2816. doi: 10.3892/mmr.2017.8198. Epub 2017 Dec 6. Mol Med Rep. 2018. PMID: 29257243 Free PMC article.
-
Unbalanced X;Y translocations carrying SRY in prenatal settings: Clinical, molecular, and cytogenetic analysis of three cases.Prenat Diagn. 2024 May;44(5):580-585. doi: 10.1002/pd.6520. Epub 2024 Jan 10. Prenat Diagn. 2024. PMID: 38204192
-
True hermaphroditism with ambiguous genitalia due to a complicated mosaic karyotype: clinical features, cytogenetic findings, and literature review.Am J Med Genet A. 2003 Jan 30;116A(3):300-3. doi: 10.1002/ajmg.a.10869. Am J Med Genet A. 2003. PMID: 12503111 Review.
-
Skewed X-chromosome inactivation pattern in SRY positive XX maleness: a case report and review of literature.Ann Genet. 2003 Jan-Mar;46(1):11-8. doi: 10.1016/s0003-3995(03)00011-x. Ann Genet. 2003. PMID: 12818524 Review.
References
-
- Cohn, R. , Scherer S., and Hamosh A.. 2020. Thompson & Thompson Genetics in Medicine. 9th ed. (Philadelphia, PA: Elsevier, 2020).
-
- Delot, E. C. , and Vilain E. J.. 2003. [Updated 2022]. “Nonsyndromic 46,XX Testicular Disorders/Differences of Sex Development.” In GeneReviews ((R)), edited by Adam M. P., Feldman J., and Mirzaa G. M., et.al, (Seattle, WA: University of Washington, 2003). https://www.ncbi.nlm.nih.gov/pubmed/20301589. - PubMed
-
- Domenice, S. , Batista R. L., Arnhold Ij P., Sircili M. H., Costa E. M. F., and Mendonca B. B.. 2000. “46,XY Differences of Sexual Development.” In Endotext, edited by Feingold K. R., Anawalt B., and Blackman M. R., et.al, (South Dartmouth, MA: MDText.com, Inc., 2000). https://www.ncbi.nlm.nih.gov/pubmed/25905393.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

