ER export via SURF4 uses diverse mechanisms of both client and coat engagement
- PMID: 39531033
- PMCID: PMC11557686
- DOI: 10.1083/jcb.202406103
ER export via SURF4 uses diverse mechanisms of both client and coat engagement
Abstract
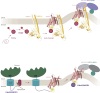
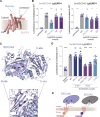
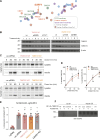
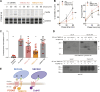
Protein secretion is an essential process that drives cell growth and communication. Enrichment of soluble secretory proteins into ER-derived transport carriers occurs via transmembrane cargo receptors that connect lumenal cargo to the cytosolic COPII coat. Here, we find that the cargo receptor, SURF4, recruits different SEC24 cargo adaptor paralogs of the COPII coat to export different cargoes. The secreted protease, PCSK9, requires both SURF4 and a co-receptor, TMED10, for export via SEC24A. In contrast, secretion of Cab45 and NUCB1 requires SEC24C/D. We further show that ER export signals of Cab45 and NUCB1 bind co-translationally to SURF4 via a lumenal pocket, contrasting prevailing models of receptor engagement only upon protein folding/maturation. Bioinformatics analyses suggest that strong SURF4-binding motifs are features of proteases, receptor-binding ligands, and Ca2+-binding proteins. We propose that certain classes of proteins are fast-tracked for rapid export to protect the health of the ER lumen.
© 2024 MRC Laboratory of Molecular Biology.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

