In utero human cytomegalovirus infection expands NK-like FcγRIII+CD8+ T cells that mediate Fc antibody functions
- PMID: 39531313
- PMCID: PMC11684805
- DOI: 10.1172/JCI181342
In utero human cytomegalovirus infection expands NK-like FcγRIII+CD8+ T cells that mediate Fc antibody functions
Abstract
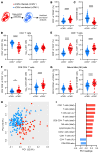
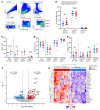
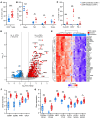
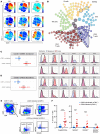
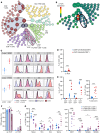
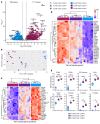
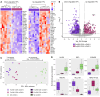
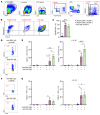
Human cytomegalovirus (HCMV) profoundly impacts host T and NK cells across the lifespan, yet how this common congenital infection modulates developing fetal immune cell compartments remains underexplored. Using cord blood from neonates with and without congenital HCMV (cCMV) infection, we identify an expansion of Fcγ receptor III-expressing (FcγRIII-expressing) CD8+ T cells following HCMV exposure in utero. Most FcγRIII+CD8+ T cells express the canonical αβ T cell receptor (TCR), but a proportion express noncanonical γδ TCR. FcγRIII+CD8+ T cells are highly differentiated and have increased expression of NK cell markers and cytolytic molecules. Transcriptional analysis reveals FcγRIII+CD8+ T cells upregulate T-bet and downregulate BCL11B, known transcription factors that govern T/NK cell fate. We show that FcγRIII+CD8+ T cells mediate antibody-dependent IFN-γ production and degranulation against IgG-opsonized target cells, similar to NK cell antibody-dependent cellular cytotoxicity (ADCC). FcγRIII+CD8+ T cell Fc effector functions were further enhanced by IL-15, as has been observed in neonatal NK cells. Our study reveals that FcγRIII+CD8+ T cells elicited in utero by HCMV infection can execute Fc-mediated effector functions bridging cellular and humoral immunity and may be a promising target for antibody-based therapeutics and vaccination in early life.
Keywords: Immunoglobulins; Immunology; Infectious disease; NK cells; T cells.
Figures
Update of
-
In utero human cytomegalovirus infection expands NK cell-like FcγRIII-expressing CD8+ T cells that mediate antibody-dependent functions.medRxiv [Preprint]. 2023 Sep 11:2023.09.08.23295279. doi: 10.1101/2023.09.08.23295279. medRxiv. 2023. Update in: J Clin Invest. 2024 Nov 12;135(1):e181342. doi: 10.1172/JCI181342. PMID: 37745390 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

