NSUN5 Facilitates Hepatocellular Carcinoma Progression by Increasing SMAD3 Expression
- PMID: 39531371
- PMCID: PMC11727281
- DOI: 10.1002/advs.202404083
NSUN5 Facilitates Hepatocellular Carcinoma Progression by Increasing SMAD3 Expression
Abstract
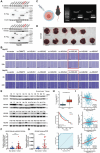
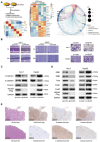
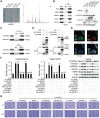
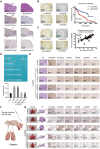
Hepatocellular carcinoma (HCC) is characterized by frequent intrahepatic and distant metastases, resulting in a poor prognosis for patients. Epithelial-mesenchymal transition (EMT) plays a pivotal role in this process. However, the expression of NOP2/Sun RNA methyltransferase 5 (NSUN5) in HCC and its role in mediating EMT remain poorly understood. In this study, clinicopathological analyses are conducted across multiple independent HCC cohorts and induced tumor formation in Nsun5-knockout mice. The findings reveal an upregulation of NSUN5 expression in tumor tissues; conversely, the absence of Nsun5 hinders the malignant progression of HCC, indicating that NSUN5 may serve as a significant oncogene in HCC. Furthermore, elevated levels of NSUN5 enhance EMT processes within HCC cells. NSUN5-knockout cells exhibit reduced invasion and migration capabilities under both in vivo and in vitro conditions, while overexpression of NSUN5 yields opposing effects. Mechanistically, high levels of NSUN5 promote the enrichment of trimethylated histone H3 at lysine 4 (H3K4me3) at the promoter region of SMAD3 through recruitment of the WDR5, thereby facilitating HCC metastasis via SMAD3-mediated EMT pathways. Collectively, this study identifies NSUN5 as a novel driver of metastasis in HCC and provides a theoretical foundation for potential therapeutic strategies against this malignancy.
Keywords: EMT; NSUN5; SMAD3; hepatocellular carcinoma (HCC); metastasis.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
- TZKY20220102/The Youth Fund of Taizhou People's Hospital Affiliated to Nanjing Medical University
- QDJJ202106/Scientific research start-up fund of Taizhou People's Hospital
- TS202308/Taizhou Society Development Project, Jiangsu, China
- H2023030/General Project of Jiangsu Provincial Health Commission
- 82372755/National Natural Science Foundation of China
- 82372746/National Natural Science Foundation of China
- 2022YQ028/Shanghai Municipal Health Commission, Health Youth Talent Project
- 20Y21902300/Shanghai Medical Innovation Research Project of "Science and Technology Innovation Action Plan"
- 2408085Y040/Natural Science Foundation of Anhui Province
- 2022zhyx-C21/Research Fund of Anhui Institute of translational medicine
- 2022AH030115/Outstanding Youth Scientific Research Projects in colleges and universities of Anhui Province
- SHDC2020CR4050/Shanghai Hospital Development Center
- SHDC12020123/Shanghai Hospital Development Center
- 21MC1930500/Shanghai Clinical Research Center of Traditional Chinese Medicine Oncology,Science and Technology Commission of Shanghai Municipality
- Qing LAN project
LinkOut - more resources
Full Text Sources
Medical
