HMGCS1 variants cause rigid spine syndrome amenable to mevalonic acid treatment in an animal model
- PMID: 39531736
- PMCID: PMC12073982
- DOI: 10.1093/brain/awae371
HMGCS1 variants cause rigid spine syndrome amenable to mevalonic acid treatment in an animal model
Abstract
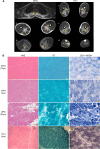
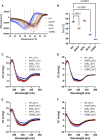
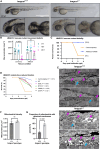
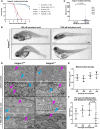
Rigid spine syndrome is a rare childhood-onset myopathy characterized by slowly progressive or non-progressive scoliosis, neck and spine contractures, hypotonia and respiratory insufficiency. Biallelic variants in SELENON account for most cases of rigid spine syndrome, however, the underlying genetic cause in some patients remains unexplained. We used exome and genome sequencing to investigate the genetic basis of rigid spine syndrome in patients without a genetic diagnosis. In five patients from four unrelated families, we identified biallelic variants in HMGCS1 (3-hydroxy-3-methylglutaryl-coenzyme A synthase). These included six missense variants and one frameshift variant distributed throughout HMGCS1. All patients presented with spinal rigidity primarily affecting the cervical and dorso-lumbar regions, scoliosis and respiratory insufficiency. Creatine kinase levels were variably elevated. The clinical course worsened with intercurrent disease or certain drugs in some patients; one patient died from respiratory failure following infection. Muscle biopsies revealed irregularities in oxidative enzyme staining with occasional internal nuclei and rimmed vacuoles. HMGCS1 encodes a critical enzyme of the mevalonate pathway and has not yet been associated with disease. Notably, biallelic hypomorphic variants in downstream enzymes including HMGCR and GGPS1 are associated with muscular dystrophy resembling our cohort's presentation. Analyses of recombinant human HMGCS1 protein and four variants (p.S447P, p.Q29L, p.M70T, p.C268S) showed that all mutants maintained their dimerization state. Three of the four mutants exhibited reduced thermal stability, and two mutants showed subtle changes in enzymatic activity compared to the wildtype. Hmgcs1 mutant zebrafish displayed severe early defects, including immobility at 2 days and death by Day 3 post-fertilisation and were rescued by HMGCS1 mRNA. We demonstrate that the four variants tested (S447P, Q29L, M70T and C268S) have reduced function compared to wild-type HMGCS1 in zebrafish rescue assays. Additionally, we demonstrate the potential for mevalonic acid supplementation to reduce phenotypic severity in mutant zebrafish. Overall, our analyses suggest that these missense variants in HMGCS1 act through a hypomorphic mechanism. Here, we report an additional component of the mevalonate pathway associated with disease and suggest biallelic variants in HMGCS1 should be considered in patients presenting with an unresolved rigid spine myopathy phenotype. Additionally, we highlight mevalonoic acid supplementation as a potential treatment for patients with HMGCS1-related disease.
Keywords: HMGCS1; enzymopathy; mevalonate pathway; neuromuscular disease; rigid spine myopathy.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Guarantors of Brain. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
The authors report no competing interests.
Figures



References
-
- Ravenscroft G, Davis MR, Lamont P, Forrest A, Laing NG. New era in genetics of early-onset muscle disease: Breakthroughs and challenges. Semin Cell Dev Biol. 2017;64:160–170. - PubMed
-
- Kazamel M, Milone M. Congenital myopathy with a novel SELN missense mutation and the challenge to differentiate it from congenital muscular dystrophy. J Clin Neurosci. 2019;62:238–239. - PubMed
-
- Tordjman M, Dabaj I, Laforet P, et al. Muscular MRI-based algorithm to differentiate inherited myopathies presenting with spinal rigidity. Eur Radiol. 2018;28:5293–5303. - PubMed
MeSH terms
Substances
Grants and funding
- UM1 HG008900/HG/NHGRI NIH HHS/United States
- Intramural Research Grant
- R01 HG009141/HG/NHGRI NIH HHS/United States
- 2020-224274/HL/NHLBI NIH HHS/United States
- R01HG009141/NH/NIH HHS/United States
- Australian NHMRC
- Silicon Valley Community Foundation
- 22ek0109490h0003/Japan Agency for Medical Research and Development
- Chan Zuckerberg Initiative DAF
- Valley Community Foundation
- DAF2019-199278/Chan Zuckerberg Initiative
- Broad Institute of MIT and Harvard Center for Mendelian Genomics
- APP2007769/Australian Government Research Training Program
- DAF2021-225399/NIH National Institute of Neurological Disorders and Stroke
- Neurological and Psychiatric Disorders of NCNP
- EY/NEI NIH HHS/United States
- U01 HG0011755/HL/NHLBI NIH HHS/United States
- UM1HG008900/NH/NIH HHS/United States
- APP1117510/Australian Government Research Training Program
- APP2009732/Australian Government Research Training Program
- APP2002640/NHMRC Ideas Grant
- U01 HG011755/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

