Divergent roles of RIPK3 and MLKL in high-fat diet-induced obesity and MAFLD in mice
- PMID: 39532538
- PMCID: PMC11557689
- DOI: 10.26508/lsa.202302446
Divergent roles of RIPK3 and MLKL in high-fat diet-induced obesity and MAFLD in mice
Abstract
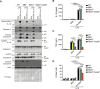
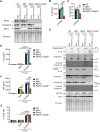
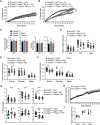
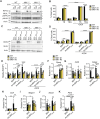
Cell death frequently occurs in the pathogenesis of obesity and metabolic dysfunction-associated fatty liver disease (MAFLD). However, the exact contribution of core cell death machinery to disease manifestations remains ill-defined. Here, we show via the direct comparison of mice genetically deficient in the essential necroptotic regulators, receptor-interacting protein kinase-3 (RIPK3) and mixed lineage kinase domain-like (MLKL), as well as mice lacking apoptotic caspase-8 in myeloid cells combined with RIPK3 loss, that RIPK3/caspase-8 signaling regulates macrophage inflammatory responses and drives adipose tissue inflammation and MAFLD upon high-fat diet feeding. In contrast, MLKL, divergent to RIPK3, contributes to both obesity and MAFLD in a manner largely independent of inflammation. We also uncover that MLKL regulates the expression of molecules involved in lipid uptake, transport, and metabolism, and congruent with this, we discover a shift in the hepatic lipidome upon MLKL deletion. Collectively, these findings highlight MLKL as an attractive therapeutic target to combat the growing obesity pandemic and metabolic disease.
© 2024 Tye et al.
Conflict of interest statement
JM Hildebrand, JM Murphy, and AL Samson contribute to, and KE Lawlor has consulted for, a project developing necroptosis inhibitors with Anaxis Pharma Pty Ltd. All other authors have no competing interests to declare.
Figures

















References
-
- Afonso MB, Rodrigues PM, Mateus-Pinheiro M, Simão AL, Gaspar MM, Majdi A, Arretxe E, Alonso C, Santos-Laso A, Jimenez-Agüero R, et al. (2021) RIPK3 acts as a lipid metabolism regulator contributing to inflammation and carcinogenesis in non-alcoholic fatty liver disease. Gut 70: 2359–2372. 10.1136/gutjnl-2020-321767 - DOI - PMC - PubMed
-
- Allam R, Lawlor KE, Yu EC, Mildenhall AL, Moujalled DM, Lewis RS, Ke F, Mason KD, White MJ, Stacey KJ, et al. (2014) Mitochondrial apoptosis is dispensable for NLRP3 inflammasome activation but non-apoptotic caspase-8 is required for inflammasome priming. EMBO Rep 15: 982–990. 10.15252/embr.201438463 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
