Hepatic SerpinA1 improves energy and glucose metabolism through regulation of preadipocyte proliferation and UCP1 expression
- PMID: 39532838
- PMCID: PMC11557585
- DOI: 10.1038/s41467-024-53835-9
Hepatic SerpinA1 improves energy and glucose metabolism through regulation of preadipocyte proliferation and UCP1 expression
Abstract
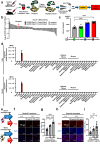
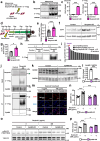
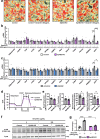
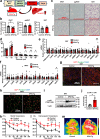
Lipodystrophy and obesity are associated with insulin resistance and metabolic syndrome accompanied by fat tissue dysregulation. Here, we show that serine protease inhibitor A1 (SerpinA1) expression in the liver is increased during recovery from lipodystrophy caused by the adipocyte-specific loss of insulin signaling in mice. SerpinA1 induces the proliferation of white and brown preadipocytes and increases the expression of uncoupling protein 1 (UCP1) to promote mitochondrial activation in mature white and brown adipocytes. Liver-specific SerpinA1 transgenic mice exhibit increased browning of adipose tissues, leading to increased energy expenditure, reduced adiposity and improved glucose tolerance. Conversely, SerpinA1 knockout mice exhibit decreased adipocyte mitochondrial function, impaired thermogenesis, obesity, and systemic insulin resistance. SerpinA1 forms a complex with the Eph receptor B2 and regulates its downstream signaling in adipocytes. These results demonstrate that SerpinA1 is an important hepatokine that improves obesity, energy expenditure and glucose metabolism by promoting preadipocyte proliferation and activating mitochondrial UCP1 expression in adipocytes.
© 2024. The Author(s).
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

