HSF-1 promotes longevity through ubiquilin-1-dependent mitochondrial network remodelling
- PMID: 39532882
- PMCID: PMC11557981
- DOI: 10.1038/s41467-024-54136-x
HSF-1 promotes longevity through ubiquilin-1-dependent mitochondrial network remodelling
Abstract
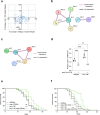
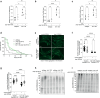
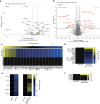
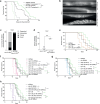
Increased activity of the heat shock factor, HSF-1, suppresses proteotoxicity and enhances longevity. However, the precise mechanisms by which HSF-1 promotes lifespan are unclear. Using an RNAi screen, we identify ubiquilin-1 (ubql-1) as an essential mediator of lifespan extension in worms overexpressing hsf-1. We find that hsf-1 overexpression leads to transcriptional downregulation of all components of the CDC-48-UFD-1-NPL-4 complex, which is central to both endoplasmic reticulum and mitochondria associated protein degradation, and that this is complemented by UBQL-1-dependent turnover of NPL-4.1. As a consequence, mitochondrial network dynamics are altered, leading to increased lifespan. Together, our data establish that HSF-1 mediates lifespan extension through mitochondrial network adaptations that occur in response to down-tuning of components associated with organellar protein degradation pathways.
© 2024. The Author(s).
Conflict of interest statement
Figures
References
-
- Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. Hallmarks of aging: An expanding universe. Cell186, 243–278 (2023). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

