Mechanics-activated fibroblasts promote pulmonary group 2 innate lymphoid cell plasticity propelling silicosis progression
- PMID: 39532893
- PMCID: PMC11557922
- DOI: 10.1038/s41467-024-54174-5
Mechanics-activated fibroblasts promote pulmonary group 2 innate lymphoid cell plasticity propelling silicosis progression
Abstract
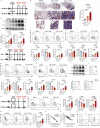
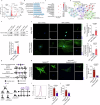
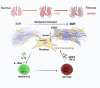
Crystalline silica (CS) particle exposure leads to silicosis which is characterized as progressive fibrosis. Fibroblasts are vital effector cells in fibrogenesis. Emerging studies have identified immune sentinel roles for fibroblasts in chronic disease, while their immune-modulatory roles in silicosis remain unclear. Herein, we show that group 2 innate lymphoid cell (ILC2) conversion to ILC1s is closely involved in silicosis progression, which is mediated by activated fibroblasts via interleukin (IL)-18. Mechanistically, Notch3 signaling in mechanics-activated fibroblasts modulates IL-18 production via caspase 1 activity. The mouse-specific Notch3 knockout in fibroblasts retards pulmonary fibrosis progression that is linked to attenuated ILC conversion. Our results indicate that activated fibroblasts in silicotic lungs are regulators of ILC2-ILC1 conversion, associated with silicosis progression via the Notch3-IL-18 signaling axis. This finding broadens our understanding of immune-modulatory mechanisms in silicosis, and indicates potential therapeutic targets for lung fibrotic diseases.
© 2024. The Author(s).
Conflict of interest statement
Figures




References
-
- Cavalin, C. et al. Beyond silicosis, is the world failing on silica hazards? Lancet Respir. Med.7, 649–650 (2019). - PubMed
-
- Hoy, R. F. & Chambers, D. C. Silica-related diseases in the modern world. Allergy75, 2805–2817 (2020). - PubMed
-
- Si, S. et al. The Australian work exposures study: prevalence of occupational exposure to respirable crystalline silica. Ann. Occup. Hyg.60, 631–637 (2016). - PubMed
-
- Davidson, S. et al. Fibroblasts as immune regulators in infection, inflammation and cancer. Nat. Rev. Immunol.21, 704–717 (2021). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

