Optimisation of cell fate determination for cultivated muscle differentiation
- PMID: 39532984
- PMCID: PMC11557827
- DOI: 10.1038/s42003-024-07201-6
Optimisation of cell fate determination for cultivated muscle differentiation
Abstract
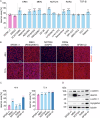
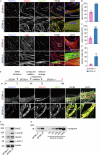
Production of cultivated meat requires defined medium formulations for the robust differentiation of myogenic cells into mature skeletal muscle fibres in vitro. Although these formulations can drive myogenic differentiation levels comparable to serum-starvation-based protocols, the resulting cultures are often heterogeneous, with a significant proportion of cells not participating in myofusion, limiting maturation of the muscle. To address this problem, we employed RNA sequencing to analyse heterogeneity in differentiating bovine satellite cells at single-nucleus resolution, identifying distinct cellular subpopulations including proliferative cells that fail to exit the cell cycle and quiescent 'reserve cells' that do not commit to myogenic differentiation. Our findings indicate that the MEK/ERK, NOTCH, and RXR pathways are active during the initial stages of myogenic cell fate determination, and by targeting these pathways, we can promote cell cycle exit while reducing reserve cell formation. This optimised medium formulation consistently yields fusion indices close to 100% in 2D culture. Furthermore, we demonstrate that these conditions enhance myotube formation and actomyosin accumulation in 3D bovine skeletal muscle constructs, providing proof of principle for the generation of highly differentiated cultivated muscle with excellent mimicry to traditional muscle.
© 2024. The Author(s).
Conflict of interest statement
Figures



References
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

