Genome-wide copy number variation association study in anorexia nervosa
- PMID: 39533101
- PMCID: PMC12014356
- DOI: 10.1038/s41380-024-02811-2
Genome-wide copy number variation association study in anorexia nervosa
Abstract
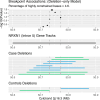
This study represents the first large-scale investigation of rare (<1% population frequency) copy number variants (CNVs) in anorexia nervosa (AN). Large, rare CNVs are reported to be causally associated with anthropometric traits, neurodevelopmental disorders, and schizophrenia, yet their role in the genetic basis of AN is unclear. Using genome-wide association study (GWAS) array data from the Anorexia Nervosa Genetics Initiative (ANGI), which included 7414 AN case and 5044 controls, we investigated the association of 67 well-established syndromic CNVs and 178 pleiotropic disease-risk dosage-sensitive CNVs with AN. To identify novel CNV regions (CNVRs) that increase the risk of AN, we conducted genome-wide association studies with a focus on rare CNV-breakpoints (CNV-GWAS). We found no net enrichment of rare CNVs, either deletions or duplications, in AN, and none of the well-established syndromic or pleiotropic CNVs had a significant association with AN status. However, the CNV-GWAS found 21 nominally associated CNVRs that contribute to AN risk, covering protein-coding genes implicated in synaptic function, metabolic/mitochondrial factors, and lipid characteristics, like the CD36 (7q21.11) gene, which transports long-chain fatty acids into cells. CNVRs intersecting genes previously related to neurodevelopmental traits include deletions of NRXN1 intron 5 (2p16.3), IMMP2L (7q31.1), and PTPRD (9p23). Overall, given that our study is well powered to detect the CNV burden level reported for schizophrenia, we can conclude that rare CNVs have a limited role in the etiology of AN, as reported for bipolar disorder. Our nominal associations for the 21 discovered CNVRs are consistent with AN being a metabo-psychiatric trait, as demonstrated by the common genetic architecture of AN, and we provide association results to allow for replication in future research.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: CMB reports receiving royalties from Pearson Education, Inc. The authors have no conflicts of interest to declare.
Figures

References
-
- Arcelus J, Mitchell AJ, Wales J, Nielsen S. Mortality rates in patients with anorexia nervosa and other eating disorders. A meta-analysis of 36 studies. Arch Gen Psychiatry. 2011;68:724–31. - PubMed
MeSH terms
Grants and funding
- R01 MH136149/MH/NIMH NIH HHS/United States
- R01 MH118278/MH/NIMH NIH HHS/United States
- R01MH136148; R56MH129437; R01MH120170; R01MH124871; R01MH118278; R01MH124871/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- R01 MH120170/MH/NIMH NIH HHS/United States
- R276-2018-4581/Lundbeckfonden (Lundbeck Foundation)
LinkOut - more resources
Full Text Sources

