CircRNA profiling of skeletal muscle satellite cells in goats reveals circTGFβ2 promotes myoblast differentiation
- PMID: 39533172
- PMCID: PMC11555921
- DOI: 10.1186/s12864-024-11008-4
CircRNA profiling of skeletal muscle satellite cells in goats reveals circTGFβ2 promotes myoblast differentiation
Abstract
Background: Circular RNAs (circRNAs) function as essential regulatory elements with pivotal roles in various biological processes. However, their expression profiles and functional regulation during the differentiation of goat myoblasts have not been thoroughly explored. This study conducts an analysis of circRNA expression profiles during the proliferation phase (cultured in growth medium, GM) and differentiation phase (cultured in differentiation medium, DM1/DM5) of skeletal muscle satellite cells (MuSCs) in goats.
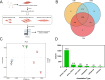
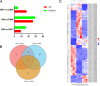
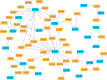
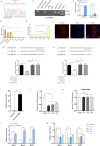
Results: A total of 2,094 circRNAs were identified, among which 84 were differentially expressed as determined by pairwise comparisons across three distinct groups. Validation of the expression levels of six randomly selected circRNAs was performed using reverse transcription PCR (RT-PCR) and quantitative RT-PCR (qRT-PCR), with confirmation of their back-splicing junction sites. Enrichment analysis of the host genes associated with differentially expressed circRNAs (DEcircRNAs) indicated significant involvement in biological processes such as muscle contraction, muscle hypertrophy, and muscle tissue development. Additionally, these host genes were implicated in key signaling pathways, including Hippo, TGF-beta, and MAPK pathways. Subsequently, employing Cytoscape, we developed a circRNA-miRNA interaction network to elucidate the complex regulatory mechanisms underlying goat muscle development, encompassing 21 circRNAs and 47 miRNAs. Functional assays demonstrated that circTGFβ2 enhances myogenic differentiation in goats, potentially through a miRNA sponge mechanism.
Conclusion: In conclusion, we identified the genome-wide expression profiles of circRNAs in goat MuSCs during both proliferation and differentiation phases, and established that circTGFβ2 plays a role in the regulation of myogenesis. This study offers a significant resource for the advanced exploration of the biological functions and mechanisms of circRNAs in the myogenesis of goats.
Keywords: Differential expression; Differentiation; Goat; circRNA sequencing; circTGFβ2.
© 2024. The Author(s).
Conflict of interest statement
Figures


Similar articles
-
Role of circPAPD7 in regulating proliferation and differentiation of goat skeletal muscle satellite cells.Genomics. 2024 Sep;116(5):110936. doi: 10.1016/j.ygeno.2024.110936. Epub 2024 Sep 14. Genomics. 2024. PMID: 39284386
-
Transcriptome Analysis Reveals the Profile of Long Non-Coding RNAs during Myogenic Differentiation in Goats.Int J Mol Sci. 2023 Mar 28;24(7):6370. doi: 10.3390/ijms24076370. Int J Mol Sci. 2023. PMID: 37047345 Free PMC article.
-
CircRNA Profiling of Skeletal Muscle in Two Pig Breeds Reveals CircIGF1R Regulates Myoblast Differentiation via miR-16.Int J Mol Sci. 2023 Feb 14;24(4):3779. doi: 10.3390/ijms24043779. Int J Mol Sci. 2023. PMID: 36835196 Free PMC article.
-
Circular RNAs and host genes act synergistically in regulating cellular processes and functions in skeletal myogenesis.Gene. 2025 Mar 10;940:149189. doi: 10.1016/j.gene.2024.149189. Epub 2024 Dec 24. Gene. 2025. PMID: 39724991 Review.
-
Circular RNAs in myogenesis.Biochim Biophys Acta Gene Regul Mech. 2020 Apr;1863(4):194372. doi: 10.1016/j.bbagrm.2019.02.011. Epub 2019 Apr 1. Biochim Biophys Acta Gene Regul Mech. 2020. PMID: 30946990 Free PMC article. Review.
Cited by
-
Advancements in Stem Cell Applications for Livestock Research: A Review.Vet Sci. 2025 Apr 23;12(5):397. doi: 10.3390/vetsci12050397. Vet Sci. 2025. PMID: 40431490 Free PMC article. Review.
-
Epigenetic Regulation of Chromatin Functions by MicroRNAs and Long Noncoding RNAs and Implications in Human Diseases.Biomedicines. 2025 Mar 16;13(3):725. doi: 10.3390/biomedicines13030725. Biomedicines. 2025. PMID: 40149701 Free PMC article. Review.
References
-
- Rehfeldt C, Fiedler I, Stickland NC. Number and size of muscle fibres in relation to meat production. CABI. 2004:1–38.
-
- Du M, Wang B, Fu X, Yang Q, Zhu M-J. Fetal programming in meat production. Meat Sci. 2015;109:40–7. - PubMed
-
- Kopantseva EE, Belyavsky AV. Key regulators of skeletal myogenesis. Mol Biol. 2016;50(2):169–92. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

