The PI3K-AKT-mTOR axis persists as a therapeutic dependency in KRASG12D-driven non-small cell lung cancer
- PMID: 39533328
- PMCID: PMC11555833
- DOI: 10.1186/s12943-024-02157-x
The PI3K-AKT-mTOR axis persists as a therapeutic dependency in KRASG12D-driven non-small cell lung cancer
Abstract
Introduction: KRASG12C and KRASG12D inhibitors represent a major translational breakthrough for non-small cell lung cancer (NSCLC) and cancer in general by directly targeting its most mutated oncoprotein. However, resistance to these small molecules has highlighted the need for rational combination partners necessitating a critical understanding of signaling downstream of KRAS mutant isoforms.
Methods: We contrasted tumor development between KrasG12C and KrasG12D genetically engineered mouse models (GEMMs). To corroborate findings and determine mutant subtype-specific dependencies, isogenic models of KrasG12C and KrasG12D initiation and adaptation were profiled by RNA sequencing. We also employed cell line models of established KRAS mutant NSCLC and determined therapeutic vulnerabilities through pharmacological inhibition. We analysed differences in survival outcomes for patients affected by advanced KRASG12C or KRASG12D-mutant NSCLC.
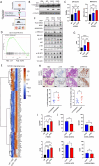
Results: KRASG12D exhibited higher potency in vivo, manifesting as more rapid lung tumor formation and reduced survival of KRASG12D GEMMs compared to KRASG12C. This increased potency, recapitulated in an isogenic initiation model, was associated with enhanced PI3K-AKT-mTOR signaling. However, KRASG12C oncogenicity and downstream pathway activation were comparable with KRASG12D at later stages of tumorigenesis in vitro and in vivo, consistent with similar clinical outcomes in patients. Despite this, established KRASG12D NSCLC models depended more on the PI3K-AKT-mTOR pathway, while KRASG12C models on the MAPK pathway. Specifically, KRASG12D inhibition was enhanced by AKT inhibition in vitro and in vivo.
Conclusions: Our data highlight a unique combination treatment vulnerability and suggest that patient selection strategies for combination approaches using direct KRAS inhibitors should be i) contextualised to individual RAS mutants, and ii) tailored to their downstream signaling.
Keywords: KRAS; KRASG12D inhibition; NSCLC; PI3K-AKT-mTOR pathway.
© 2024. The Author(s).
Conflict of interest statement
Figures





References
-
- Thomas A, Liu SV, Subramaniam DS, Giaccone G. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat Rev Clin Oncol. 2015;12:511–26. - PubMed
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. - PubMed
MeSH terms
Substances
Grants and funding
- A27412/CRUK Manchester Institute (CRUK MI)
- A27412/CRUK Manchester Institute (CRUK MI)
- A27412/CRUK Manchester Institute (CRUK MI)
- A27412/CRUK Manchester Institute (CRUK MI)
- A27412/CRUK Manchester Institute (CRUK MI)
- A27412/CRUK Manchester Institute (CRUK MI)
- A27412/CRUK Manchester Institute (CRUK MI)
- A27412/CRUK Manchester Institute (CRUK MI)
- A25254/Manchester CRUK Centre Award
- A25254/Manchester CRUK Centre Award
- A25146/CRUK Manchester Experimental Cancer Medicines Centre
- A25146/CRUK Manchester Experimental Cancer Medicines Centre
- A20465/CRUK Lung Cancer Centre of Excellence
- A20465/CRUK Lung Cancer Centre of Excellence
- PID2021-124632OB-100/MINECO/FEDER
- PID2021-124632OB-100/MINECO/FEDER
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

