Mapping the evolution of liver aging research: A bibliometric analysis
- PMID: 39534417
- PMCID: PMC11551677
- DOI: 10.3748/wjg.v30.i41.4461
Mapping the evolution of liver aging research: A bibliometric analysis
Abstract
Background: With the increasing of the global aging population, healthy aging and prevention of age-related diseases have become increasingly important. The liver, a vital organ involved in metabolism, detoxification, digestion, and immunity, holds a pivotal role in the aging process of organisms. Although extensive research on liver aging has been carried out, no bibliometric analysis has been conducted to evaluate the scientific progress in this area.
Aim: To analyze basic knowledge, development trends, and current research frontiers in the field via bibliometric methods.
Methods: We conducted bibliometric analyses via a range of analytical tools including Python, the bibliometrix package in R, CiteSpace, and VOSviewer. We retrieved publication data on liver aging research from the Web of Science Core Collection Database. A scientific knowledge map was constructed to display the contributions from different authors, journals, countries, institutions, as well as patterns of co-occurrence keywords and co-cited references. Additionally, gene regulation pathways associated with liver aging were analyzed via the STRING database.
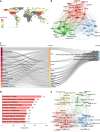
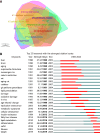
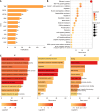
Results: We identified 4288 articles on liver aging, authored by 24034 contributors from 4092 institutions across 85 countries. Notably, the years 1991 and 2020 presented significant bursts in publication output. The United States led in terms of publications (n = 1008, 25.1%), citations (n = 55205), and international collaborations (multiple country publications = 214). Keywords such as "lipid metabolism", "fatty liver disease", "inflammation", "liver fibrosis" and "target" were prominent, highlighting the current research hotspots. Notably, the top 64 genes, each of which appeared in at least 8 articles, were involved in pathways essential for cell survival and aging, including the phosphatidylinositol 3-kinase/protein kinase B, Forkhead box O and p53 signaling pathways.
Conclusion: This study highlights key areas of liver aging and offers a comprehensive overview of research trends, as well as insights into potential value for collaborative pursuits and clinical implementations.
Keywords: Aging; Bibliometric; CiteSpace; Gene regulation; Liver; R language; VOSviewer.
©The Author(s) 2024. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: All the authors report no relevant conflicts of interest for this article.
Figures







References
-
- Ma S, Chi X, Cai Y, Ji Z, Wang S, Ren J, Liu GH. Decoding Aging Hallmarks at the Single-Cell Level. Annu Rev Biomed Data Sci. 2023;6:129–152. - PubMed
-
- Wu Z, Qu J, Zhang W, Liu GH. Stress, epigenetics, and aging: Unraveling the intricate crosstalk. Mol Cell. 2024;84:34–54. - PubMed
-
- López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. Hallmarks of aging: An expanding universe. Cell. 2023;186:243–278. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

