Exploring heterocyclic scaffolds in carbonic anhydrase inhibition: a decade of structural and therapeutic insights
- PMID: 39534850
- PMCID: PMC11555472
- DOI: 10.1039/d4ra06290f
Exploring heterocyclic scaffolds in carbonic anhydrase inhibition: a decade of structural and therapeutic insights
Abstract
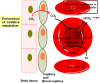
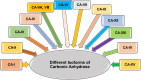
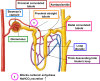
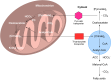
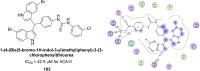
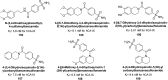
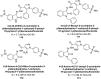
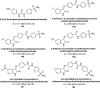
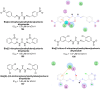
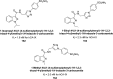
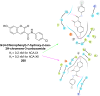
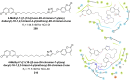
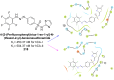
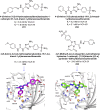
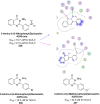
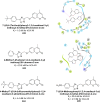
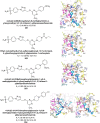
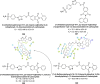
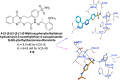
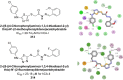
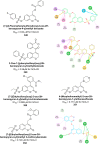
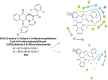
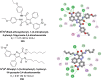
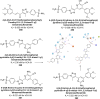
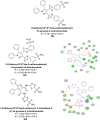
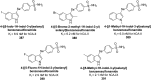
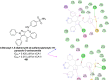
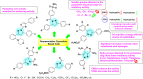
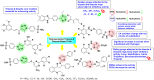
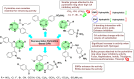
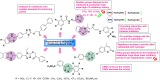
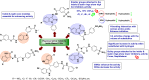
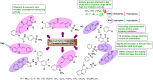
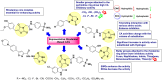
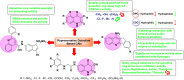
Heterocyclic compounds represent a prominent class of molecules with diverse pharmacological activities. Among their therapeutic applications, they have gained significant attention as carbonic anhydrase (CA) inhibitors, owing to their potential in the treatment of various diseases such as epilepsy, cancer and glaucoma. CA is a widely distributed zinc metalloenzyme that facilitates the reversible interconversion of carbon dioxide and bicarbonate. This reaction is essential for numerous physiological and pathological processes. In humans, CA exists in sixteen different isoforms, labeled hCA-I to hCA-XV, each distributed across various tissues and organs and involved in crucial physiological functions. Clinically utilized CA inhibitors, such as brinzolamide, dorzolamide and acetazolamide, exhibit poor selectivity, leading to undesirable side effects. A significant challenge in designing effective CA inhibitors is achieving balanced isoform selectivity, prompting the exploration of new chemotypes. This review compiles recent strategies employed by various researchers in developing CAIs across different structural classes, including pyrazoline, quinoline, imidazole, oxadiazole, pyrimidine, coumarin, chalcone, rhodanine, phthalazine, triazole, isatin, and indole. Additionally, the review summarizes structure-activity relationship (SAR) analyses, isoform selectivity evaluations, along with mechanistic and in silico investigations. Insights derived from SAR studies provide crucial directions for the rational design of next-generation heterocyclic CA inhibitors, with improved therapeutic efficacy and reduced side effects. To the best of our knowledge, for the first time, we have comprehensively summarized all known isoforms of CA in relation to various heterocyclic motifs. This review examines the use of different heterocycles as CA inhibitors, drawing on research published over the past 11 years. It offers a valuable resource for early-career researchers, encouraging further exploration of synthetic heterocycles in the development of CA inhibitors.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
Authors have no conflict of interests to declare.
Figures









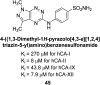



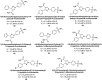
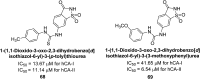

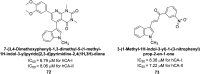



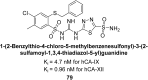

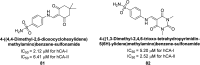
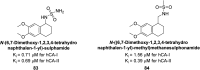

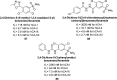
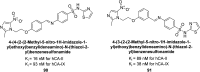








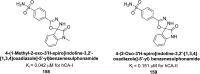








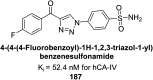
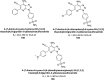

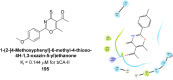


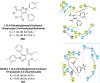
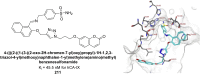









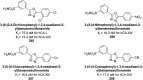
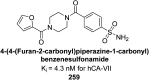
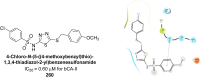







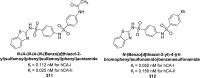










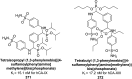








Similar articles
-
Recent advances in the medicinal chemistry of carbonic anhydrase inhibitors.Eur J Med Chem. 2021 Jan 1;209:112923. doi: 10.1016/j.ejmech.2020.112923. Epub 2020 Oct 15. Eur J Med Chem. 2021. PMID: 33121862 Review.
-
Carbonic anhydrases: Moiety appended derivatives, medicinal and pharmacological implications.Bioorg Med Chem. 2024 Nov 15;114:117933. doi: 10.1016/j.bmc.2024.117933. Epub 2024 Oct 3. Bioorg Med Chem. 2024. PMID: 39378610 Review.
-
Progress in the development of human carbonic anhydrase inhibitors and their pharmacological applications: Where are we today?Med Res Rev. 2020 Nov;40(6):2485-2565. doi: 10.1002/med.21713. Epub 2020 Jul 21. Med Res Rev. 2020. PMID: 32691504 Review.
-
Carbonic anhydrase inhibitors: synthesis and inhibition of the human carbonic anhydrase isoforms I, II, VII, IX and XII with benzene sulfonamides incorporating 4,5,6,7-tetrabromophthalimide moiety.Bioorg Med Chem. 2013 Oct 1;21(19):5973-82. doi: 10.1016/j.bmc.2013.07.044. Epub 2013 Aug 2. Bioorg Med Chem. 2013. PMID: 23965175
-
Structural study of interaction between brinzolamide and dorzolamide inhibition of human carbonic anhydrases.Bioorg Med Chem. 2013 Nov 15;21(22):7210-5. doi: 10.1016/j.bmc.2013.08.033. Epub 2013 Aug 28. Bioorg Med Chem. 2013. PMID: 24090602
Cited by
-
In Vitro Leishmanicidal Efficacy of Synthesized Arylidene Analogues of Glitazone.Drug Dev Res. 2025 Aug;86(5):e70125. doi: 10.1002/ddr.70125. Drug Dev Res. 2025. PMID: 40654248 Free PMC article.
References
-
- Qadir T. Amin A. Sharma P. K. Jeelani I. Abe H. Open Med. Chem. J. 2022;16:1–34.
-
- Kalaria P. N. Karad S. C. Raval D. K. Eur. J. Med. Chem. 2018;158:917–936. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

