Integrin β8 Facilitates Macrophage Infiltration and Polarization by Regulating CCL5 to Promote LUAD Progression
- PMID: 39535362
- PMCID: PMC11727125
- DOI: 10.1002/advs.202406865
Integrin β8 Facilitates Macrophage Infiltration and Polarization by Regulating CCL5 to Promote LUAD Progression
Abstract
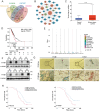
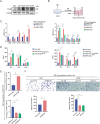
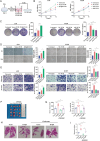
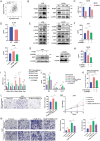
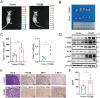
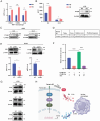
The tumor microenvironment (TME) influences cancer progression and metastasis. Integrin β8 (ITGβ8), a member of the integrin family, is upregulated in various cancers. In this study, it is determined as a key factor that mediates the interaction between lung adenocarcinoma (LUAD) cells and macrophages. Increased expression levels of ITGβ8 are associated with increased numbers of CD163+ macrophages and poor prognosis in LUAD patients. The overexpression of ITGβ8 in LUAD cells promotes the polarization of THP-1 macrophages toward the M2 phenotype. In contrast, TCM (conditioned medium from the co-culture system) from THP-1 macrophages and ITGβ8-overexpressing A549 cells promoted the proliferation and invasion of A549 cells. Mechanistically, chemokine (C-C motif) ligand 5 (CCL5) plays an important role in mediating ITGβ8-induced macrophage polarization, and the phosphoinositide 3-kinase (PI3K)/AKT serine/threonine kinase (AKT)/interferon regulatory factor 9 (IRF9) pathway is involved in this process. Moreover, interleukin 8 (IL8) and interleukin 10 (IL10) produced by M2-like macrophages regulate the expression of ITGβ8 in LUAD cells through the spi-1 proto-oncogene (SPI1). This study elucidates the feedback mechanism of ITGβ8 between LUAD cells and macrophages.
Keywords: CCL5; ITGβ8; LUAD; TME; macrophage.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Bray F., Laversanne M., Sung H., Ferlay J., Siegel R., Soerjomataram I., Jemal A., CA Cancer J. Clin. 2024, 74, 229. - PubMed
-
- Leiter A., Veluswamy R., Wisnivesky J., Nat. Rev. Clin. Oncol. 2023, 20, 624. - PubMed
-
- Romain R., Christian B., Jorge G., Diane D., Marie C., Dieu N., Catherine S., Wolf H., Charles A., Nasser K., Miriam M., Sacha G., Am J. Respir. Crit. Care Med. 2014, 191, 90.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
