Inactivation of Pseudovirus Expressing the D614G Spike Protein Mutation using Nitric Oxide-Plasma Activated Water
- PMID: 39535372
- PMCID: PMC11672301
- DOI: 10.1002/advs.202411515
Inactivation of Pseudovirus Expressing the D614G Spike Protein Mutation using Nitric Oxide-Plasma Activated Water
Abstract
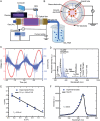
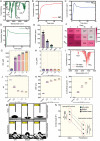
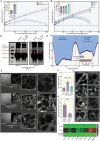
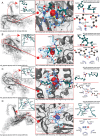
Variants of concern (VOCs) of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) exhibit high infectivity due to mutations, particularly in the spike protein, that facilitate enhanced binding of virus to human angiotensin-converting enzyme 2 (hACE2). The D614G mutation, situated in S1-domain, promotes the open conformation of spike protein, augmenting its interaction with hACE2. Activated water neutralizes pathogens by damaging biological molecules; however, its effect on mutated SARS-CoV-2 or VOCs requires further exploration. Here, the efficacy of nitric oxide (NOx)-plasma activated water (PAW) in inhibiting infections by SARS-CoV-2 pseudovirus expressing D614G-mutated spike protein is investigated, which serves as a model for mutated SARS-CoV-2. Results demonstrated high prevalence of D614G mutation in SARS-CoV-2 and its VOCs. NOx-PAW is non-toxic to cells at high concentration, inhibiting infection by 71%. Moreover, NOx-PAW induced structural changes in S1-domain of spike protein, reducing its binding affinity and lowering clathrin-mediated endocytosis-related gene expression. Additionally, in silico analysis revealed NOx species in NOx-PAW played key role in impairing S1-domain function of the mutated SARS-CoV-2 pseudovirus by interacting directly with it. Collectively, these findings reveal the potent inactivation ability of PAW against mutated SARS-CoV-2 and suggest its potential application in combating emerging variants of SARS-CoV-2 and other viral threats.
Keywords: SARS‐CoV‐2 pseudovirus D614G spike mutant; mutated virus inactivation; nitric oxides; plasma activated water; variants of concern.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
V367F Mutation in SARS-CoV-2 Spike RBD Emerging during the Early Transmission Phase Enhances Viral Infectivity through Increased Human ACE2 Receptor Binding Affinity.J Virol. 2021 Jul 26;95(16):e0061721. doi: 10.1128/JVI.00617-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34105996 Free PMC article.
-
The Spike-Stabilizing D614G Mutation Interacts with S1/S2 Cleavage Site Mutations To Promote the Infectious Potential of SARS-CoV-2 Variants.J Virol. 2022 Oct 12;96(19):e0130122. doi: 10.1128/jvi.01301-22. Epub 2022 Sep 19. J Virol. 2022. PMID: 36121299 Free PMC article.
-
The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types.Elife. 2021 Feb 11;10:e65365. doi: 10.7554/eLife.65365. Elife. 2021. PMID: 33570490 Free PMC article.
-
Proteolytic activation of SARS-CoV-2 spike protein.Microbiol Immunol. 2022 Jan;66(1):15-23. doi: 10.1111/1348-0421.12945. Epub 2021 Oct 12. Microbiol Immunol. 2022. PMID: 34561887 Free PMC article. Review.
-
Structural and functional insights into the spike protein mutations of emerging SARS-CoV-2 variants.Cell Mol Life Sci. 2021 Dec;78(24):7967-7989. doi: 10.1007/s00018-021-04008-0. Epub 2021 Nov 3. Cell Mol Life Sci. 2021. PMID: 34731254 Free PMC article. Review.
References
-
- COVID – Coronavirus Statistics – Worldometer, https://www.worldometers.info/coronavirus/, (accessed: February 2024).
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
