Epidemiologic and Genomic Characterization of an Outbreak of Rift Valley Fever Among Humans and Dairy Cattle in Northern Tanzania
- PMID: 39535803
- PMCID: PMC12069657
- DOI: 10.1093/infdis/jiae562
Epidemiologic and Genomic Characterization of an Outbreak of Rift Valley Fever Among Humans and Dairy Cattle in Northern Tanzania
Abstract
Background: A periurban outbreak of Rift Valley fever virus (RVFV) among dairy cattle from May through August 2018 in northern Tanzania was detected through testing samples from prospective livestock abortion surveillance. We sought to identify concurrent human infections, their phylogeny, and epidemiologic characteristics in a cohort of febrile patients enrolled from 2016 to 2019 at hospitals serving the epizootic area.
Methods: From September 2016 through May 2019, we conducted a prospective cohort study that enrolled febrile patients hospitalized at 2 hospitals in Moshi, Tanzania. Archived serum, plasma, or whole blood samples were retrospectively tested for RVFV by PCR. Human samples positive for RVFV were sequenced and compared to RVFV sequences obtained from cattle through a prospective livestock abortion study. Phylogenetic analysis was performed on complete RVFV genomes.
Results: Among 656 human participants, we detected RVFV RNA in 4 (0.6%), including 1 death with hepatic necrosis and other end-organ damage at autopsy. Humans infected with RVFV were enrolled from June through August 2018, and all resided in or near urban areas. Phylogenetic analysis of human and cattle RVFV sequences demonstrated that most clustered to lineage B, a lineage previously described in East Africa. A lineage E strain clustering with lineages in Angola was also identified in cattle.
Conclusions: We provide evidence that an apparently small RVFV outbreak among dairy cattle in northern Tanzania was associated with concurrent severe and fatal infections among humans. Our findings highlight the unidentified scale and diversity of interepizootic RVFV transmission, including near and within an urban area.
Keywords: Africa; Crimean-Congo hemorrhagic fever; Rift Valley fever; Tanzania; zoonoses.
Published by Oxford University Press on behalf of Infectious Diseases Society of America 2024.
Conflict of interest statement
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Figures
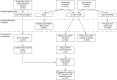



References
-
- Daubney R, Hudson JR, Garnham PC. Enzootic hepatitis or Rift Valley fever. An undescribed virus disease of sheep cattle and man from East Africa. J Pathol Bacteriol 1931; 34:545–79.
-
- Gerdes GH. Rift Valley fever. Rev Sci Tech 2004; 23:613–23. - PubMed
MeSH terms
Substances
Grants and funding
- 5T32AI007392/US National Institute of Health
- R83537/University of Edinburgh
- K08 AI182480/AI/NIAID NIH HHS/United States
- U01 AI062563/AI/NIAID NIH HHS/United States
- BB/R020280/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- K23 AI116869/AI/NIAID NIH HHS/United States
- CC999999/ImCDC/Intramural CDC HHS/United States
- D43TW009337/NIH Fogarty International Center
- R01TW009237/US National Institute of Health
- R01 AI121378/AI/NIAID NIH HHS/United States
- R01 TW009237/TW/FIC NIH HHS/United States
- T32 AI007392/AI/NIAID NIH HHS/United States
- D43TW009337/US National Institute of Health
- D43 TW009337/TW/FIC NIH HHS/United States
LinkOut - more resources
Full Text Sources

