Distribution of Clostridioides difficile ribotypes and sequence types across humans, animals and food in 13 European countries
- PMID: 39535868
- PMCID: PMC11610360
- DOI: 10.1080/22221751.2024.2427804
Distribution of Clostridioides difficile ribotypes and sequence types across humans, animals and food in 13 European countries
Abstract
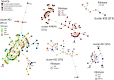
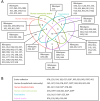
Clostridioides difficile is a One Health pathogen found in humans, animals, and the environment, with food representing a potential transmission route. One Health studies are often limited to a single country or selected reservoirs and ribotypes. This study provides a varied and accessible collection of C. difficile isolates and sequencing data derived from human, animal, and food sources across 13 European countries. A total of 441 strains (human hospital- and community-associated cases n = 280, animal n = 96, food n = 65) were analysed by ribotyping, toxinotyping and whole-genome sequencing (WGS). We detected 83 sequence types (STs), with ST11 (n = 80 isolates) and ST1 (n = 54 isolates) being the most represented. Several STs included strains originating from all source combinations. Further genomic analysis confirmed close genetic relatedness in some of the STs. Additionally, the genomic analysis identified 10 strains from cryptic clades (C-I to C-III) and 4 of them were mono-toxigenic possessing only a variant form of tcdA gene. Amongst 106 ribotypes, 10 were shared between all 3 sources and 68 were source-specific. Some ribotypes were only found at the intersection of human and food source (RT023, RT027), or between human and animal source (RT009, RT045, RT046). C. difficile ribotypes and STs in Europe were diverse. In this collection, some ribotypes showed potential association with food or animal transmission routes. C. difficile strains from divergent clades CI-III, currently emerging in the human population, were rare and mostly food-associated.Trial registration: ClinicalTrials.gov identifier: NCT03503474.
Keywords: One Health; animals; community-associated; cryptic clades; epidemiology; food; hospital-associated; transmission.
Conflict of interest statement
ND and BS are employees of the GSK group of companies. ND receives restricted shares from GSK as part of her employee remuneration. BM, ESA, PC are employees of the bioMérieux company. MR, VT, SJ, VV, GD, KD, MW report no conflict of interest related to this work. KD and MW are supported in part by the National Institute for Health and Care Research (NIHR) Leeds Biomedical Research Centre (BRC) (NIHR203331). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care.
Figures



References
-
- Williamson CHD, Stone NE, Nunnally AE, et al. . Identification of novel, cryptic Clostridioides species isolates from environmental samples collected from diverse geographical locations. Microb Genom. 2022 Feb;8(2):000742. doi:10.1099/mgen.0.000742. PMID: 35166655; PMCID: PMC8942030. - DOI - PMC - PubMed
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
