Creeping Stem 1 regulates directional auxin transport for lodging resistance in soybean
- PMID: 39535932
- PMCID: PMC11772330
- DOI: 10.1111/pbi.14503
Creeping Stem 1 regulates directional auxin transport for lodging resistance in soybean
Abstract
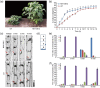
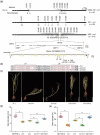
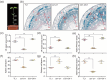
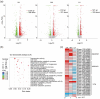
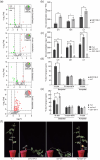
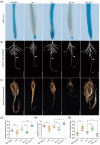
Soybean, a staple crop on a global scale, frequently encounters challenges due to lodging under high planting densities, which results in significant yield losses. Despite extensive research, the fundamental genetic mechanisms governing lodging resistance in soybeans remain elusive. In this study, we identify and characterize the Creeping Stem 1 (CS1) gene, which plays a crucial role in conferring lodging resistance in soybeans. The CS1 gene encodes a HEAT-repeat protein that modulates hypocotyl gravitropism by regulating amyloplast sedimentation. Functional analysis reveals that the loss of CS1 activity disrupts polar auxin transport, vascular bundle development and the biosynthesis of cellulose and lignin, ultimately leading to premature lodging and aberrant root development. Conversely, increasing CS1 expression significantly enhances lodging resistance and improves yield under conditions of high planting density. Our findings shed light on the genetic mechanisms that underlie lodging resistance in soybeans and highlight the potential of CS1 as a valuable target for genetic engineering to improve crop lodging resistance and yield.
Keywords: amyloplast sedimentation; gravitropism; lodging resistance; polar auxin transport; soybean.
© 2024 The Author(s). Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures

Similar articles
-
LRM3 positively regulates stem lodging resistance by degradating MYB6 transcriptional repressor in soybean.Plant Biotechnol J. 2025 Jul;23(7):2978-2993. doi: 10.1111/pbi.70124. Epub 2025 May 7. Plant Biotechnol J. 2025. PMID: 40333576 Free PMC article.
-
Soybean hypocotyl elongation is regulated by a MYB33-SWEET11/21-GA2ox8c module involving long-distance sucrose transport.Plant Biotechnol J. 2024 Oct;22(10):2859-2872. doi: 10.1111/pbi.14409. Epub 2024 Jun 11. Plant Biotechnol J. 2024. PMID: 38861663 Free PMC article.
-
Mutation of OsPIN1b by CRISPR/Cas9 Reveals a Role for Auxin Transport in Modulating Rice Architecture and Root Gravitropism.Int J Mol Sci. 2022 Aug 11;23(16):8965. doi: 10.3390/ijms23168965. Int J Mol Sci. 2022. PMID: 36012245 Free PMC article.
-
Root gravitropism: a complex response to a simple stimulus?Trends Plant Sci. 1999 Oct;4(10):407-12. doi: 10.1016/s1360-1385(99)01472-7. Trends Plant Sci. 1999. PMID: 10498965 Review.
-
Auxin regulated gene expression and gravitropism in plants.ASGSB Bull. 1995 Oct;8(2):39-45. ASGSB Bull. 1995. PMID: 11538549 Review.
Cited by
-
LRM3 positively regulates stem lodging resistance by degradating MYB6 transcriptional repressor in soybean.Plant Biotechnol J. 2025 Jul;23(7):2978-2993. doi: 10.1111/pbi.70124. Epub 2025 May 7. Plant Biotechnol J. 2025. PMID: 40333576 Free PMC article.
References
-
- Antwi‐Boasiako, A. (2017) Screening of soybean (Glycine max (L.) Merrill) genotypes for resistance to lodging and pod shattering. Int. J. Agron. Agric. Res. 10, 1–8.
-
- Baker, C.J. , Sterling, M. and Berry, P. (2014) A generalised model of crop lodging. J. Theor. Biol. 363, 1–12. - PubMed
-
- Baldwin, K.L. , Strohm, A.K. and Masson, P.H. (2013) Gravity sensing and signal transduction in vascular plant primary roots. Am. J. Bot. 100, 126–142. - PubMed
-
- Chen, H. , Yang, Z. , Chen, L. , Zhang, C. , Yuan, S. , Zhang, X. , Qiu, D. et al. (2017) Combining QTL and candidate gene analysis with phenotypic model to unravel the relationship between lodging and related traits in soybean. Mol. Breed. 37, 1–14. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

