ACE2-independent sarbecovirus cell entry can be supported by TMPRSS2-related enzymes and can reduce sensitivity to antibody-mediated neutralization
- PMID: 39536058
- PMCID: PMC11559990
- DOI: 10.1371/journal.ppat.1012653
ACE2-independent sarbecovirus cell entry can be supported by TMPRSS2-related enzymes and can reduce sensitivity to antibody-mediated neutralization
Abstract
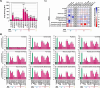
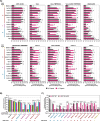
The COVID-19 pandemic, caused by SARS-CoV-2, demonstrated that zoonotic transmission of animal sarbecoviruses threatens human health but the determinants of transmission are incompletely understood. Here, we show that most spike (S) proteins of horseshoe bat and Malayan pangolin sarbecoviruses employ ACE2 for entry, with human and raccoon dog ACE2 exhibiting broad receptor activity. The insertion of a multibasic cleavage site into the S proteins increased entry into human lung cells driven by most S proteins tested, suggesting that acquisition of a multibasic cleavage site might increase infectivity of diverse animal sarbecoviruses for the human respiratory tract. In contrast, two bat sarbecovirus S proteins drove cell entry in an ACE2-independent, trypsin-dependent fashion and several ACE2-dependent S proteins could switch to the ACE2-independent entry pathway when exposed to trypsin. Several TMPRSS2-related cellular proteases but not the insertion of a multibasic cleavage site into the S protein allowed for ACE2-independent entry in the absence of trypsin and may support viral spread in the respiratory tract. Finally, the pan-sarbecovirus antibody S2H97 enhanced cell entry driven by two S proteins and this effect was reversed by trypsin while trypsin protected entry driven by a third S protein from neutralization by S2H97. Similarly, plasma from quadruple vaccinated individuals neutralized entry driven by all S proteins studied, and availability of the ACE2-independent, trypsin-dependent pathway reduced neutralization sensitivity. In sum, our study reports a pathway for entry into human cells that is ACE2-independent, can be supported by TMPRSS2-related proteases and may be associated with antibody evasion.
Copyright: © 2024 Zhang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
SP and MH conducted contract research (testing of vaccinee plasma for neutralizing activity against SARS-CoV-2) for Valneva unrelated to this work. GMNB served as advisor for Moderna and SP served as advisor for BioNTech, unrelated to this work. MSW. received funding from Sartorius AG (Göttingen, Germany) from GRIFOLS SA (Barcelona, Spain), Sphingotec (Henningsdorf, Germany), Inflammatix (Sunnyvale, CA, USA) and the German Research Foundation (Bonn, Germany) unrelated to this work. MSW is in the advisory board of Amomed (Wien, Austria) and Gilead Science Inc. (Foster City, CA, USA). All other authors declare no competing interests.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

