The vacuolar K+/H+ exchangers and calmodulin-like CML18 constitute a pH-sensing module that regulates K+ status in Arabidopsis
- PMID: 39536104
- PMCID: PMC11559620
- DOI: 10.1126/sciadv.adp7658
The vacuolar K+/H+ exchangers and calmodulin-like CML18 constitute a pH-sensing module that regulates K+ status in Arabidopsis
Abstract
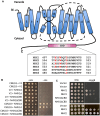
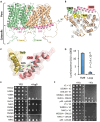
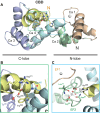
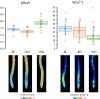
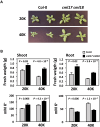
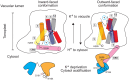
Shifts in cytosolic pH have been recognized as key signaling events and mounting evidence supports the interdependence between H+ and Ca2+ signaling in eukaryotic cells. Among the cellular pH-stats, K+/H+ exchange at various membranes is paramount in plant cells. Vacuolar K+/H+ exchangers of the NHX (Na+,K+/H+ exchanger) family control luminal pH and, together with K+ and H+ transporters at the plasma membrane, have been suggested to also regulate cytoplasmic pH. We show the regulation of vacuolar K+/H+ exchange by cytoplasmic pH and the calmodulin-like protein CML18 in Arabidopsis. The crystal structure and physicochemical properties of CML18 indicate that this protein senses pH shifts. Interaction of CML18 with tonoplast exchangers NHX1 and NHX2 was favored at acidic pH, a physiological condition elicited by K+ starvation in Arabidopsis roots, whereas excess K+ produced cytoplasmic alkalinization and CML18 dissociation. These results imply that the pH-responsive NHX-CML18 module is an essential component of the cellular K+- and pH-stats.
Figures




References
-
- Pardo J. M., Cubero B., Leidi E. O., Quintero F. J., Alkali cation exchangers: Roles in cellular homeostasis and stress tolerance. J. Exp. Bot. 57, 1181–1199 (2006). - PubMed
-
- Venema K., Quintero F. J., Pardo J. M., Donaire J. P., The Arabidopsis Na+/H+ exchanger AtNHX1 catalyzes low affinity Na+ and K+ transport in reconstituted liposomes. J. Biol. Chem. 277, 2413–2418 (2002). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

