Toxic small alarmone synthetase FaRel2 inhibits translation by pyrophosphorylating tRNAGly and tRNAThr
- PMID: 39536105
- PMCID: PMC11559606
- DOI: 10.1126/sciadv.adr9624
Toxic small alarmone synthetase FaRel2 inhibits translation by pyrophosphorylating tRNAGly and tRNAThr
Abstract
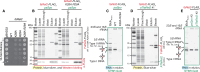
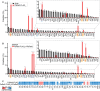
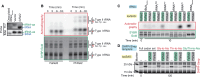
Translation-targeting toxic small alarmone synthetases (toxSAS) are effectors of bacterial toxin-antitoxin systems that pyrophosphorylate the 3'-CCA end of transfer RNA (tRNA) to prevent aminoacylation. toxSAS are implicated in antiphage immunity: Phage detection triggers the toxSAS activity to shut down viral production. We show that the toxSAS FaRel2 inspects the tRNA acceptor stem to specifically select tRNAGly and tRNAThr. The first, second, fourth, and fifth base pairs of the stem act as the specificity determinants. We show that the toxSASs PhRel2 and CapRelSJ46 differ in tRNA specificity from FaRel2 and rationalize this through structural modeling: While the universal 3'-CCA end slots into a highly conserved CCA recognition groove, the acceptor stem recognition region is variable across toxSAS diversity. As phages use tRNA isoacceptors to overcome tRNA-targeting defenses, we hypothesize that highly evolvable modular tRNA recognition allows for the escape of viral countermeasures through tRNA substrate specificity switching.
Figures


Update of
-
Toxic Small Alarmone Synthetase FaRel2 inhibits translation by pyrophosphorylating tRNAGly and tRNAThr.bioRxiv [Preprint]. 2024 Jul 5:2024.07.05.602228. doi: 10.1101/2024.07.05.602228. bioRxiv. 2024. Update in: Sci Adv. 2024 Nov 15;10(46):eadr9624. doi: 10.1126/sciadv.adr9624. PMID: 39005314 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

