APEX1 Polymorphisms Affect Acute Myeloid Leukemia Risk, and Its Expression Is Involved in Cell Proliferation and Differentiation
- PMID: 39536468
- PMCID: PMC11885693
- DOI: 10.1111/ijlh.14401
APEX1 Polymorphisms Affect Acute Myeloid Leukemia Risk, and Its Expression Is Involved in Cell Proliferation and Differentiation
Abstract
Introduction: The link between DNA repair gene polymorphisms and cancer susceptibility has gained significant attention. Thus, we investigated the impact of base excision repair (BER) gene polymorphisms on acute myeloid leukemia (AML) risk and pathogenesis.
Methods: In total, 106 patients with AML and 191 healthy controls were included in the study, wherein polymorphisms in four BER genes (APEX1, MUTYH, OGG1, and XRCC1) were examined.
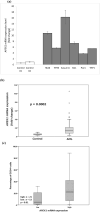
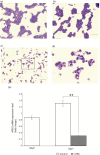
Results: Notably, the APEX1-656 T>G polymorphism exhibited a significant association with AML risk in the recessive (TT vs. TG + GG) (p = 0.046) and co-dominant models (TT vs. GG) (p = 0.02). Assessing APEX1 expression levels, APEX1 expression was elevated in the bone marrow of patients with AML compared with that in controls (p = 0.02). Subsequently, we compared the percentages of CD34+ cells between the APEX1 high or low expression groups, revealing a significant difference (high vs. low = 29.9% vs. 11.5%, p = 0.01). Additionally, we observed reduced APEX1 expression in HL60 cells differentiated with all-trans retinoic acid (p < 0.001). We hypothesized that APEX1 expression could correlate with stemness and analyzed its expression in stem and differentiated cells.
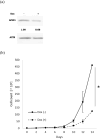
Conclusions: In the GSE48558 dataset, AML cells and normal CD34+ cells expressed APEX1 at higher levels than did granulocytes (p < 0.01). Functional experiments revealed that APEX1 knockdown led to a reduction in AML cell proliferation. These findings indicated that APEX1 polymorphisms were a potential risk factor for AML and highlighted the important role of APEX1 in regulating AML cell differentiation and proliferation.
Keywords: acute myeloid leukemia; base excision repair; differentiation; expression; polymorphism.
© 2024 The Author(s). International Journal of Laboratory Hematology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing or financial interest in this study.
Figures
References
-
- Kirtonia A., Pandya G., Sethi G., Pandey A. K., Das B. C., and Garg M., “A Comprehensive Review of Genetic Alterations and Molecular Targeted Therapies for the Implementation of Personalized Medicine in Acute Myeloid Leukemia,” Journal of Molecular Medicine 98, no. 8 (2020): 1069–1091, 10.1007/s00109-020-01944-5. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

