A p38 MAPK-ROS axis fuels proliferation stress and DNA damage during CRISPR-Cas9 gene editing in hematopoietic stem and progenitor cells
- PMID: 39536752
- PMCID: PMC11604517
- DOI: 10.1016/j.xcrm.2024.101823
A p38 MAPK-ROS axis fuels proliferation stress and DNA damage during CRISPR-Cas9 gene editing in hematopoietic stem and progenitor cells
Abstract
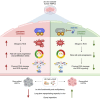
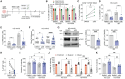
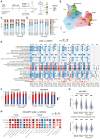
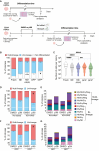
Ex vivo activation is a prerequisite to reaching adequate levels of gene editing by homology-directed repair (HDR) for hematopoietic stem and progenitor cell (HSPC)-based clinical applications. Here, we show that shortening culture time mitigates the p53-mediated DNA damage response to CRISPR-Cas9-induced DNA double-strand breaks, enhancing the reconstitution capacity of edited HSPCs. However, this results in lower HDR efficiency, rendering ex vivo culture necessary yet detrimental. Mechanistically, ex vivo activation triggers a multi-step process initiated by p38 mitogen-activated protein kinase (MAPK) phosphorylation, which generates mitogenic reactive oxygen species (ROS), promoting fast cell-cycle progression and subsequent proliferation-induced DNA damage. Thus, p38 inhibition before gene editing delays G1/S transition and expands transcriptionally defined HSCs, ultimately endowing edited cells with superior multi-lineage differentiation, persistence throughout serial transplantation, enhanced polyclonal repertoire, and better-preserved genome integrity. Our data identify proliferative stress as a driver of HSPC dysfunction with fundamental implications for designing more effective and safer gene correction strategies for clinical applications.
Keywords: CRISPR-Cas9; DNA damage; DNA damage response; cell cycle; clonal output; differentiation; gene editing; hematopoietic stem cells; p38 MAPK-ROS; proliferative stress; single-cell analyses.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.D.M., L.d.V., F.M., A.C., L.N., and S.F. are inventors of patents on applications of gene editing in HSPCs owned and managed by the San Raffaele Scientific Institute and the Telethon Foundation. L.N. is a founder and quota holder of GeneSpire.
Figures



References
-
- Ferrari S., Valeri E., Conti A., Scala S., Aprile A., Di Micco R., Kajaste-Rudnitski A., Montini E., Ferrari G., Aiuti A., Naldini L. Genetic engineering meets hematopoietic stem cell biology for next-generation gene therapy. Cell Stem Cell. 2023;30:549–570. - PubMed
-
- Doudna J.A., Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346 - PubMed
-
- Porteus M.H. A New Class of Medicines through DNA Editing. N. Engl. J. Med. 2019;380:947–959. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

