The mitochondrial mRNA-stabilizing protein SLIRP regulates skeletal muscle mitochondrial structure and respiration by exercise-recoverable mechanisms
- PMID: 39537626
- PMCID: PMC11561311
- DOI: 10.1038/s41467-024-54183-4
The mitochondrial mRNA-stabilizing protein SLIRP regulates skeletal muscle mitochondrial structure and respiration by exercise-recoverable mechanisms
Abstract
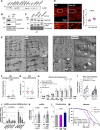
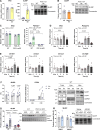
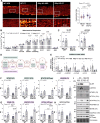
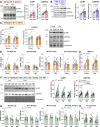
Decline in mitochondrial function is linked to decreased muscle mass and strength in conditions like sarcopenia and type 2 diabetes. Despite therapeutic opportunities, there is limited and equivocal data regarding molecular cues controlling muscle mitochondrial plasticity. Here we uncovered that the mitochondrial mRNA-stabilizing protein SLIRP, in complex with LRPPRC, is a PGC-1α target that regulates mitochondrial structure, respiration, and mtDNA-encoded-mRNA pools in skeletal muscle. Exercise training effectively counteracts mitochondrial defects caused by genetically-induced LRPPRC/SLIRP loss, despite sustained low mtDNA-encoded-mRNA pools, by increasing mitoribosome translation capacity and mitochondrial quality control. In humans, exercise training robustly increases muscle SLIRP and LRPPRC protein across exercise modalities and sexes, yet less prominently in individuals with type 2 diabetes. SLIRP muscle loss reduces Drosophila lifespan. Our data points to a mechanism of post-transcriptional mitochondrial regulation in muscle via mitochondrial mRNA stabilization, offering insights into how exercise enhances mitoribosome capacity and mitochondrial quality control to alleviate defects.
© 2024. The Author(s).
Conflict of interest statement
Figures


References
-
- Janssen, I., Heymsfield, S. B. & Ross, R. Low relative skeletal muscle mass (sarcopenia) in older persons is associated with functional impairment and physical disability. J. Am. Geriatr. Soc.50, 889–896 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NNF16OC0023418/Novo Nordisk Fonden (Novo Nordisk Foundation)
- NNF18OC0032082/Novo Nordisk Fonden (Novo Nordisk Foundation)
- NNF20OC0063577/Novo Nordisk Fonden (Novo Nordisk Foundation)
- 801199/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- R380-2021-1451/Lundbeckfonden (Lundbeck Foundation)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

