Genome assembly at chromosome scale with telomere ends for Pearlspot, Etroplus suratensis
- PMID: 39537670
- PMCID: PMC11560961
- DOI: 10.1038/s41597-024-04096-0
Genome assembly at chromosome scale with telomere ends for Pearlspot, Etroplus suratensis
Abstract
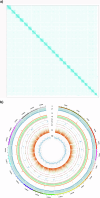
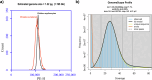
The pearlspot, Etroplus suratensis is a climate resilient cichlid fish that exhibits unusual adaptation to salinity. The fish is able to complete full life cycle in diverse salinity habitats ranging from fresh water to marine environments. High-quality primary and phased genome assemblies were generated for pearlspot fish using PacBio HiFi and Arima HiC sequencing technologies, for the first time. The primary assembly is highly contiguous with contig N50 length of 36 Mb. The final assembly is of 1.247 Gb with N50 length of 51.57 Mb and 98% of the genome length anchored to 24 chromosomes. The genome was assessed to be 99.9% complete based on BUSCO evaluation and was predicted to contain 52.96% repeat elements. We have predicted 27,192 protein encoding genes, of which 21,580 were functionally annotated. The genome offers an invaluable resource to understand adaptation of pearlspot fish to diverse salinity habitats.
© 2024. The Author(s).
Conflict of interest statement
Figures




Similar articles
-
A telomere-to-telomere chromosome-scale genome assembly of glass catfish (Kryptopterus vitreolus).Sci Data. 2025 Mar 23;12(1):483. doi: 10.1038/s41597-025-04841-z. Sci Data. 2025. PMID: 40122884 Free PMC article.
-
A complete telomere-to-telomere chromosome-level genome assembly of X-ray tetra (Pristella maxillaris).Sci Data. 2025 Mar 24;12(1):496. doi: 10.1038/s41597-025-04824-0. Sci Data. 2025. PMID: 40128533 Free PMC article.
-
Sea lice, Caligus rotundigenitalis infestations and its management in pond cultured pearlspot, Etroplus suratensis in Gujarat: a case study.J Parasit Dis. 2016 Jun;40(2):565-7. doi: 10.1007/s12639-014-0539-y. Epub 2014 Aug 31. J Parasit Dis. 2016. PMID: 27413343 Free PMC article.
-
A natural outbreak of infectious spleen and kidney necrosis virus threatens wild pearlspot, Etroplus suratensis in Peechi Dam in the Western Ghats biodiversity hotspot, India.Transbound Emerg Dis. 2022 Sep;69(5):e1595-e1605. doi: 10.1111/tbed.14494. Epub 2022 Mar 8. Transbound Emerg Dis. 2022. PMID: 35235241
-
Resistance of pearlspot larvae, Etroplus suratensis, to redspotted grouper nervous necrosis virus by immersion challenge.J Fish Dis. 2019 Feb;42(2):249-256. doi: 10.1111/jfd.12930. Epub 2018 Nov 29. J Fish Dis. 2019. PMID: 30488969
Cited by
-
Chromosome-level genome assembly with telomeric repeats at scaffold ends for Rhabdosargus sarba.Sci Data. 2025 Jul 11;12(1):1202. doi: 10.1038/s41597-025-05324-x. Sci Data. 2025. PMID: 40645974 Free PMC article.
References
-
- Schliewen, U. K., Tautz, D. & Pääbo, S. Sympatric speciation suggested by monophyly of crater lake cichlids. Nature368, 629–632 (1994). - PubMed
-
- Ronco, F. et al. Drivers and dynamics of a massive adaptive radiation in cichlid fishes. Nature589, 76–81 (2021). - PubMed
-
- Kocher, T. D. Adaptive evolution and explosive speciation: the cichlid fish model. Nat Rev Genet5, 288–298 (2004). - PubMed
-
- Ward, J. A. & Wyman, R. L. Ethology and ecology of cichlid fishes of the genus Etroplus in Sri Lanka: preliminary findings. Environ Biol Fishes2, 137–145 (1977).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

