Characterization of variably protease-sensitive prionopathy by capillary electrophoresis
- PMID: 39537719
- PMCID: PMC11561330
- DOI: 10.1038/s41598-024-79217-1
Characterization of variably protease-sensitive prionopathy by capillary electrophoresis
Abstract
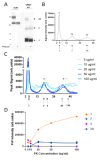
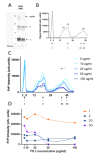
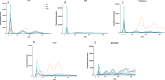
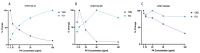
Variably Protease Sensitive Prionopathy (VPSPr) is a rare human prion disease that, like Creutzfeldt-Jakob disease (CJD), results in the deposition of abnormally folded prion protein aggregates in the brain and is ultimately fatal. Neuropathology and clinical features of VPSPr are heterogeneous. However, the key discriminating feature is the relative sensitivity of the pathological prion protein to proteinase digestion compared to that typically seen in other human prion cases. Three major fragments of 23, 17 and 7 kDa are characteristic of the disease following digestion with proteinase K. We recently reported the utility of the highly adaptive and reproducible ProteinSimple™ capillary electrophoresis (CE) system to perform protein separation of PK digested prion protein in CJD. Consequently, we explored capillary-based electrophoresis (CE) technology as a sensitive method to detect and characterize VPSPr in a cohort of 29 cases. The unique 7 kDa fragment has high intensity, particularly in cases with the codon 129 VV genotype, but can be missed by regular Western blotting due to the small size. However, this fragment is readily detected by CE in all cases. In addition, the flexibility of CE produced highly reproducible, semi-quantitative data for determining relative proteinase K sensitivity and epitope mapping of representative cases from each codon 129 genotype (VV, MV and MM).
© 2024. Crown.
Conflict of interest statement
Figures




References
-
- Sitammagari, K. K. & Masood, W. Creutzfeldt Jakob Disease, In: StatPearls, StatPearls Publishing, Treasure Island (FL), http://www.ncbi.nlm.nih.gov/books/NBK507860/ (Accessed 25 July 2023). - PubMed
-
- Hill, A. F. et al. Molecular classification of sporadic Creutzfeldt-Jakob disease. Brain J. Neurol.126, 1333–1346. 10.1093/brain/awg125 (2003). - PubMed
-
- Gambetti, P., Kong, Q., Zou, W., Parchi, P. & Chen, S. G. Sporadic and familial CJD: classification and characterisation. Br. Med. Bull.66, 213–239. 10.1093/bmb/66.1.213 (2003). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

