Hepatocellular senescence induces multi-organ senescence and dysfunction via TGFβ
- PMID: 39537753
- PMCID: PMC11628396
- DOI: 10.1038/s41556-024-01543-3
Hepatocellular senescence induces multi-organ senescence and dysfunction via TGFβ
Abstract
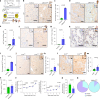
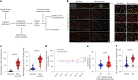
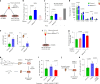
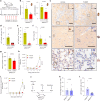
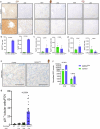
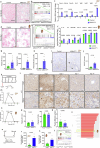
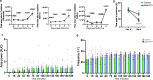
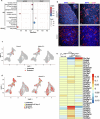
Cellular senescence is not only associated with ageing but also impacts physiological and pathological processes, such as embryonic development and wound healing. Factors secreted by senescent cells affect their microenvironment and can induce spreading of senescence locally. Acute severe liver disease is associated with hepatocyte senescence and frequently progresses to multi-organ failure. Why the latter occurs is poorly understood. Here we demonstrate senescence development in extrahepatic organs and associated organ dysfunction in response to liver senescence using liver injury models and genetic models of hepatocyte-specific senescence. In patients with severe acute liver failure, we show that the extent of hepatocellular senescence predicts disease outcome, the need for liver transplantation and the occurrence of extrahepatic organ failure. We identify the TGFβ pathway as a critical mediator of systemic spread of senescence and demonstrate that TGFβ inhibition in vivo blocks senescence transmission to other organs, preventing liver senescence induced renal dysfunction. Our results highlight the systemic consequences of organ-specific senescence, which, independent of ageing, contributes to multi-organ dysfunction.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: S.T.B. is an employee and shareholder of AstraZeneca. L.A.B. and F.O. are directors, employees and shareholders of Fibrofind, and L.H.R. is an employee and shareholder of Fibrofind. A patent entitled ‘Agents for use in a method of treating an acute liver disease’ has been filed in relation to the work associating senescence markers with patient outcomes; patent applicants: Beatson Institute for Cancer Research, University College London, Royal Free London NHS Foundation Trust, Charité – Universitätsmedizin Berlin; inventor(s)—T.G.B., F.A., R.J. and A.Q. (application number 2309181.2). R.J. is the inventor of OPA, which has been patented by UCL and licensed to Mallinckrodt Pharma. He is also the founder of Yaqrit Limited, Hepyx Limited (spin out companies from University College London) and Cyberliver. He has research collaborations with Yaqrit Limited. The other authors do not have competing interests to declare.
Figures




References
-
- Munoz-Espin, D. & Serrano, M. Cellular senescence: from physiology to pathology. Nat. Rev. Mol. Cell Biol.15, 482–496 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- WT107492Z/Wellcome Trust (Wellcome)
- 945096/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- MR/R023026/1/MRC_/Medical Research Council/United Kingdom
- A29802/Cancer Research UK (CRUK)
- A17196/Cancer Research UK (CRUK)
- MR/X004112/1/RCUK | Medical Research Council (MRC)
- 226202/Z/22/Z/Wellcome Trust (Wellcome)
- MR/X008304/1/RCUK | MRC | Medical Research Foundation
- A31287/Cancer Research UK (CRUK)
- A25045/Cancer Research UK (CRUK)
- MR/W00089X/1/RCUK | Medical Research Council (MRC)
- WT_/Wellcome Trust/United Kingdom
- MR/K0019494/1/RCUK | Medical Research Council (MRC)
- A26813/Cancer Research UK (CRUK)
- MR/L016354/1/MRC_/Medical Research Council/United Kingdom
- PGL22/100014/Rosetrees Trust
- 23390/CRUK_/Cancer Research UK/United Kingdom
- MR/Y003365/1/RCUK | MRC | Medical Research Foundation
- MR/Y003365/1/RCUK | Medical Research Council (MRC)
- A21139/Cancer Research UK (CRUK)
- MR/K001949/1/MRC_/Medical Research Council/United Kingdom
- HPRU-2012-10076/DH | National Institute for Health Research (NIHR)
- MR/R006237/1/RCUK | MRC | Medical Research Foundation
- MR/N013166/1/RCUK | Medical Research Council (MRC)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

