Releasing a sugar brake generates sweeter tomato without yield penalty
- PMID: 39537922
- PMCID: PMC11578880
- DOI: 10.1038/s41586-024-08186-2
Releasing a sugar brake generates sweeter tomato without yield penalty
Abstract
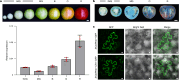
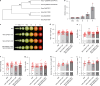
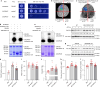
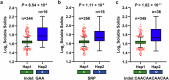
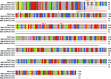
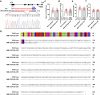
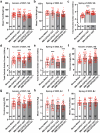
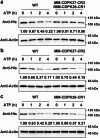
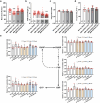
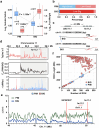
In tomato, sugar content is highly correlated with consumer preferences, with most consumers preferring sweeter fruit1-4. However, the sugar content of commercial varieties is generally low, as it is inversely correlated with fruit size, and growers prioritize yield over flavour quality5-7. Here we identified two genes, tomato (Solanum lycopersicum) calcium-dependent protein kinase 27 (SlCDPK27; also known as SlCPK27) and its paralogue SlCDPK26, that control fruit sugar content. They act as sugar brakes by phosphorylating a sucrose synthase, which promotes degradation of the sucrose synthase. Gene-edited SlCDPK27 and SlCDPK26 knockouts increased glucose and fructose contents by up to 30%, enhancing perceived sweetness without fruit weight or yield penalty. Although there are fewer, lighter seeds in the mutants, they exhibit normal germination. Together, these findings provide insight into the regulatory mechanisms controlling fruit sugar accumulation in tomato and offer opportunities to increase sugar content in large-fruited cultivars without sacrificing size and yield.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: A patent on the use of SlCDPK27 and SlCDPK26 to improve fruit sugar content has been filed by S.H., J.Z., H.L., J.C. and G.Z. (CN202211616622.8; PCT/CN2023/138699). The other authors declare no competing interests.
Figures




References
-
- Gough, C. & Hobson, G. E. A comparison of the productivity, quality, shelf-life characteristics and consumer reaction to the crop from cherry tomato plants grown at different levels of salinity. J. Hortic. Sci.65, 431–439 (1990).
-
- Kader, A. A. Flavor quality of fruits and vegetables. J. Sci. Food Agric.88, 1863–1868 (2008).
-
- Beckles, D. M. Factors affecting the postharvest soluble solids and sugar content of tomato (Solanum lycopersicum L.) fruit. Postharvest Biol. Technol.63, 129–140 (2012).
-
- Cong, B., Barrero, L. S. & Tanksley, S. D. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet.40, 800–804 (2008). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

