Impact of mixed Staphylococcus aureus-Pseudomonas aeruginosa biofilm on susceptibility to antimicrobial treatments in a 3D in vitro model
- PMID: 39538008
- PMCID: PMC11561256
- DOI: 10.1038/s41598-024-79573-y
Impact of mixed Staphylococcus aureus-Pseudomonas aeruginosa biofilm on susceptibility to antimicrobial treatments in a 3D in vitro model
Abstract
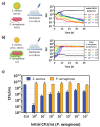
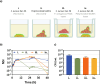
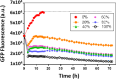
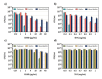
Staphylococcus aureus and Pseudomonas aeruginosa are the most common bacteria co-isolated from chronic infected wounds. Their interactions remain unclear but this coexistence is beneficial for both bacteria and may lead to resistance to antimicrobial treatments. Besides, developing an in vitro model where this coexistence is recreated remains challenging, making difficult their study. The aim of this work was to develop a reliable polymicrobial in vitro model of both species to further understand their interrelationships and the effects of different antimicrobials in coculture. In this work, bioluminescent and fluorescent bacteria were used to evaluate the activity of two antiseptics (chlorhexidine and thymol) against these bacteria planktonically grown, or when forming single and mixed biofilms. At the doses tested (0.4-1,000 mg/L), thymol showed selective antimicrobial action against S. aureus in planktonic and biofilm states, in contrast with chlorhexidine which exerted antimicrobial effects against both bacteria. Furthermore, the initial conditions for both bacteria in the co-culture determined the antimicrobial outcome, showing that P. aeruginosa impaired the proliferation and metabolism of S. aureus. Moreover, S. aureus showed an increased tolerance against antiseptic treatments when co-cultured, attributed to the formation of a thicker mixed biofilm compared to those obtained when monocultured, and also, by the reduction of S. aureus metabolic activity induced by diffusible molecules produced by P. aeruginosa. This work underlines the relevance of polymicrobial populations and their crosstalk and microenvironment in the search of disruptive and effective treatments for polymicrobial biofilms.
Keywords: Pseudomonas aeruginosa; Staphylococcus aureus; Antiseptics; Chlorhexidine; Polymicrobial biofilm; Thymol.
© 2024. The Author(s).
Conflict of interest statement
Figures




Similar articles
-
Pseudomonas aeruginosa Increases the Sensitivity of Biofilm-Grown Staphylococcus aureus to Membrane-Targeting Antiseptics and Antibiotics.mBio. 2019 Jul 30;10(4):e01501-19. doi: 10.1128/mBio.01501-19. mBio. 2019. PMID: 31363032 Free PMC article.
-
Pseudomonas aeruginosa Alters Staphylococcus aureus Sensitivity to Vancomycin in a Biofilm Model of Cystic Fibrosis Infection.mBio. 2017 Jul 18;8(4):e00873-17. doi: 10.1128/mBio.00873-17. mBio. 2017. PMID: 28720732 Free PMC article.
-
Optimal environmental and culture conditions allow the in vitro coexistence of Pseudomonas aeruginosa and Staphylococcus aureus in stable biofilms.Sci Rep. 2019 Nov 8;9(1):16284. doi: 10.1038/s41598-019-52726-0. Sci Rep. 2019. PMID: 31705015 Free PMC article.
-
Pseudomonas aeruginosa and Staphylococcus aureus communication in biofilm infections: insights through network and database construction.Crit Rev Microbiol. 2019 Sep-Nov;45(5-6):712-728. doi: 10.1080/1040841X.2019.1700209. Epub 2019 Dec 13. Crit Rev Microbiol. 2019. PMID: 31835971 Review.
-
Antimicrobial Tolerance in Cross-Kingdom Dual-Species Biofilms Formed by Fungi and Bacteria.Med Mycol J. 2024;65(3):49-57. doi: 10.3314/mmj.24.004. Med Mycol J. 2024. PMID: 39218647 Review.
References
-
- Yung, D. B. Y., Sircombe, K. J. & Pletzer, D. Friends or enemies? The complicated relationship between Pseudomonas aeruginosa and Staphylococcus aureus. Mol. Microbiol.116, 1–15 (2021). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

