MicroRNAome profiling of breast cancer unveils hsa-miR-5683 as a tumor suppressor microRNA predicting favorable clinical outcome
- PMID: 39538254
- PMCID: PMC11562357
- DOI: 10.1186/s12935-024-03550-8
MicroRNAome profiling of breast cancer unveils hsa-miR-5683 as a tumor suppressor microRNA predicting favorable clinical outcome
Abstract
Background: Breast cancer is a heterogeneous disease with diverse molecular subtypes, underscoring a better understanding of its molecular features and underlying regulatory mechanisms. Therefore, identifying novel prognostic biomarkers and therapeutic targets is crucial for advancing the current standard of care for breast cancer patients.
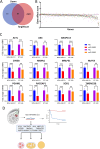
Methods: Ninety-six formalin-fixed paraffin-embedded (FFPE) breast cancer samples underwent miRNAome profiling using QIAseq microRNA library kit and sequencing on Illumina platform. Mature miRNA quantification was conducted using CLC Genomics Workbench v21.0.5, while Relapse-free survival (RFS) analysis was conducted using RStudio 2023.09.1. Gain-of-function studies were conducted using miRNA mimics, while the effects of miRNA exogenous expression on cancer hallmark were assessed using 2-dimentional (2D) proliferation assay, three-dimensional (3D) organotypic culture, and live-dead staining. TargetScan database and Ingenuity Pathway Analysis (IPA) were used for miRNA target identification.
Results: Hierarchical clustering based on miRNA expression revealed distinct patterns in relation to PAM50 classification and identified miRNAs panels associated with luminal, HER2, and basal subtypes. hsa-miR-5683 emerged as a potential prognostic biomarker, showing a favorable correlation with RFS and suppressing tumorigenicity under 2D and 3D conditions in triple-negative breast cancer (TNBC) models. Findings were further extended to the MCF7 hormone receptor positive (HR+) model. Transcriptomic profiling of hsa-miR-5683 overexpressing TNBC cells revealed its potential role in key oncogenic pathways. Integration of downregulated genes and CRISPR-Cas9 perturbational effects identified ACLY, RACGAP1, AK4, MRPL51, CYB5B, MKRN1, TMEM230, NUP54, ANAPC13, PGAM1, and SOD1 as bona fide gene targets for hsa-miR-5683.
Conclusions: Our data provides comprehensive miRNA expression atlas in breast cancer subtypes and underscores the prognostic and therapeutic significance of numerous miRNAs, including hsa-miR-5683 in TNBC. The identified gene targets unravel the intricate regulatory network in TNBC progression, suggesting promising avenues for further research and targeted therapeutic interventions.
Keywords: Breast cancer; Prognostic biomarker; Survival; Therapeutic target; microRNA.
© 2024. The Author(s).
Conflict of interest statement
Figures




References
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. Ca Cancer J Clin. 2023;73(1):17–48. - PubMed
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2021;71(3):209–49. - PubMed
-
- Alteri R, Barnes C, Burke A, Gansler T, Gapstur S, Gaudet M, Kramer J, Newman L, Niemeyer D, Richards C. Breast cancer facts & figs. Atlanta: American Cancer Society; 2013. pp. 2013–4.
-
- Bener A, Ayub H, Kakil R, Ibrahim W. Patterns of cancer incidence among the population of Qatar: a worldwide comparative study. Asian Pac J Cancer Prev. 2008;9(1):19–24. - PubMed
-
- Boyle P, Levin B. World cancer report 2008. IARC Press, International Agency for Research on Cancer; 2008.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

