Analysis of hsa_circ_0136256 as a biomarker for fibrosis in systemic sclerosis
- PMID: 39538329
- PMCID: PMC11562351
- DOI: 10.1186/s12896-024-00910-0
Analysis of hsa_circ_0136256 as a biomarker for fibrosis in systemic sclerosis
Abstract
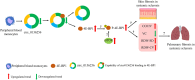
Background: Exploration of whether circRNAs in the skin of systemic sclerosis (SSc) model mice interact with 4E-BP1 protein to mediate the mTOR signaling pathway to regulate SSc fibrosis is crucial to identify homologous human circRNAs as markers to guide the diagnosis and treatment of SSc.
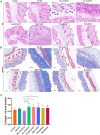
Methods: C57BL/6 mice aged 6-8 weeks and weighing approximately 20 g were subcutaneously injected with bleomycin (BLM) to establish an SSc model. High-throughput sequencing was used to screen the differentially expressed circRNA in the skin of SSc model mice and control mice. RNA immunoprecipitation and RNA pulldown confirmed the interaction between circRNA and 4E-BP1 protein. SSc model mice were treated with empty plasmid (OE-NC), overexpression plasmid of mmu_circ_0005372 (OE-circ_0005372), interference plasmid of mmu_circ_0005372 (sh-circ5372), mutant plasmid of mmu_circ_0005372 (circ5372-MT), mTOR activator (MHY1485), mTOR inhibitor (omipalisib), or JAK1/2 inhibitor (ruxolitinib). Sections of mouse skin tissue were stained with Hematoxylin and eosin and Masson's stain. The collagen volume fraction (CVF) was calculated as CVF = area of blue collagen/total area with ImageJ. The correlation between homologous human circRNAs and clinical data was analyzed.
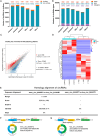
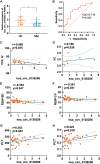
Results: Compared to the control group, 21,839 circRNAs were upregulated and 27, 946 circRNAs were downregulated in the skin tissue of mice in the SSc model group. Among them was mmu_circ_0005372, which is derived from the FZD3 gene, is closely related to fibrosis, and is involved in the mTOR signaling pathway. Hsa_circ_0136256 was identified as the homologous human circRNA of mmu_circ_0005372. RT-qPCR confirmed that the expression of mmu_circ_0005372 was significantly reduced in the skin tissue of SSc mice, and the expression of hsa_circ_0136256 was significantly reduced in the peripheral blood mononuclear cells of patients with SSc. The interaction between mmu_circ_0005372 and 4E-BP1 protein was inhibited in the skin tissue of SSc model mice. The results showed that the CVF of OE-circ_0005372 group was significantly lower than that of the sh-circ5372, circ5372-MT, and MHY1485 groups, indicating that OE-circ5372 significantly improved skin fibrosis in the SSc mice. ROC curve analysis was performed on hsa_circ_0136256 (AUC = 0.719, P = 0.035). The expression of hsa_circ_0136256 was negatively correlated with COL IV, RDW-SD, and RDW-CV, and positively correlated with VC, PLT, and PCT. The results suggested that hsa_circ_0136256 may have important roles in the clinical diagnosis of SSc.
Conclusion: Mmu_circ_0005372 and homologous human hsa_circ_0136256 may be biomarkers and therapeutic targets for SSc fibrosis.
Keywords: Fibrosis; MTOR/4E-BP1; Systemic sclerosis; circRNA.
© 2024. The Author(s).
Conflict of interest statement
Figures

Similar articles
-
Mmu_circ_0005373 and hsa_circ_0136255 participate in the pulmonary fibrosis of systemic sclerosis.Int Immunopharmacol. 2024 Sep 30;139:112690. doi: 10.1016/j.intimp.2024.112690. Epub 2024 Jul 24. Int Immunopharmacol. 2024. PMID: 39053227
-
NRF1-induced mmu_circ_0001388/hsa_circ_0029470 confers ferroptosis resistance in ischemic acute kidney injury via the miR-193b-3p/TCF4/GPX4 axis.Life Sci. 2024 Dec 1;358:123190. doi: 10.1016/j.lfs.2024.123190. Epub 2024 Oct 29. Life Sci. 2024. PMID: 39481837
-
A comprehensive evaluation of skin aging-related circular RNA expression profiles.J Clin Lab Anal. 2021 Apr;35(4):e23714. doi: 10.1002/jcla.23714. Epub 2021 Feb 3. J Clin Lab Anal. 2021. PMID: 33534927 Free PMC article.
-
The roles and mechanisms of circular RNAs related to mTOR in cancers.J Clin Lab Anal. 2022 Dec;36(12):e24783. doi: 10.1002/jcla.24783. Epub 2022 Nov 25. J Clin Lab Anal. 2022. PMID: 36426933 Free PMC article. Review.
-
Biomarkers in the management of scleroderma: an update.Curr Rheumatol Rep. 2011 Feb;13(1):4-12. doi: 10.1007/s11926-010-0140-z. Curr Rheumatol Rep. 2011. PMID: 21046295 Review.
References
-
- Sun X, Wei W, Ren J, Liang Y, Wang M, Gui Y, Xue X, Li J, Dai C. Inhibition of 4E-BP1 phosphorylation promotes tubular cell escaping from G2/M arrest and ameliorates kidney fibrosis. Cell Signal. 2019;62:109331. - PubMed
-
- Liu C, Chen L. Circular RNAs: Characterization, cellular roles, and applications. Cell. 2022;185(12):2016–34. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

