Marginal interaction test for detecting interactions between genetic marker sets and environment in genome-wide studies
- PMID: 39538414
- PMCID: PMC11708225
- DOI: 10.1093/g3journal/jkae263
Marginal interaction test for detecting interactions between genetic marker sets and environment in genome-wide studies
Abstract
As human complex diseases are influenced by the interaction between genetics and the environment, identifying gene-environment interactions (G×E) is crucial for understanding disease mechanisms and predicting risk. Developing robust quantitative tools for G×E analysis can enhance the study of complex diseases. However, many existing methods that explore G×E focus on the interplay between an environmental factor and genetic variants, exclusively for common or rare variants. In this study, we developed MAGEIT_RAN and MAGEIT_FIX to identify interactions between an environmental factor and a set of genetic markers, including both rare and common variants, based on the MinQue for Summary statistics. The genetic main effects in MAGEIT_RAN and MAGEIT_FIX are modeled as random and fixed effects, respectively. Simulation studies showed that both tests had type I error under control, with MAGEIT_RAN being the most powerful test. Applying MAGEIT to a genome-wide analysis of gene-alcohol interactions on hypertension and seated systolic blood pressure in the Multiethnic Study of Atherosclerosis revealed genes like EIF2AK2, CCNDBP1, and EPB42 influencing blood pressure through alcohol interaction. Pathway analysis identified 1 apoptosis and survival pathway involving PKR and 2 signal transduction pathways associated with hypertension and alcohol intake, demonstrating MAGEIT_RAN's ability to detect biologically relevant gene-environment interactions.
Keywords: gene–environment interaction; genome-wide study; method of moments; mixed effects model.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The authors declare no conflicts of interest.
Figures
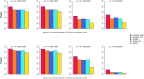


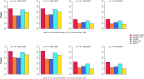
Update of
-
Detection of interactions between genetic marker sets and environment in a genome-wide study of hypertension.bioRxiv [Preprint]. 2023 May 30:2023.05.28.542666. doi: 10.1101/2023.05.28.542666. bioRxiv. 2023. Update in: G3 (Bethesda). 2025 Jan 8;15(1):jkae263. doi: 10.1093/g3journal/jkae263. PMID: 37398075 Free PMC article. Updated. Preprint.
Similar articles
-
Detection of interactions between genetic marker sets and environment in a genome-wide study of hypertension.bioRxiv [Preprint]. 2023 May 30:2023.05.28.542666. doi: 10.1101/2023.05.28.542666. bioRxiv. 2023. Update in: G3 (Bethesda). 2025 Jan 8;15(1):jkae263. doi: 10.1093/g3journal/jkae263. PMID: 37398075 Free PMC article. Updated. Preprint.
-
Using Genetic Marginal Effects to Study Gene-Environment Interactions with GWAS Data.Behav Genet. 2021 May;51(3):358-373. doi: 10.1007/s10519-021-10058-8. Epub 2021 Apr 26. Behav Genet. 2021. PMID: 33899139
-
HisCoM-G×E: Hierarchical Structural Component Analysis of Gene-Based Gene-Environment Interactions.Int J Mol Sci. 2020 Sep 14;21(18):6724. doi: 10.3390/ijms21186724. Int J Mol Sci. 2020. PMID: 32937825 Free PMC article.
-
Between candidate genes and whole genomes: time for alternative approaches in blood pressure genetics.Curr Hypertens Rep. 2012 Feb;14(1):46-61. doi: 10.1007/s11906-011-0241-8. Curr Hypertens Rep. 2012. PMID: 22161147 Free PMC article. Review.
-
A Review of the Genetics of Hypertension with a Focus on Gene-Environment Interactions.Curr Hypertens Rep. 2017 Mar;19(3):23. doi: 10.1007/s11906-017-0718-1. Curr Hypertens Rep. 2017. PMID: 28283927 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
