The Prognostic Significance of NEDD9 Expression in Human Cancers: A Systematic Review, Meta-Analysis, and Omics Exploration
- PMID: 39540210
- PMCID: PMC11561999
- DOI: 10.1177/15330338241297597
The Prognostic Significance of NEDD9 Expression in Human Cancers: A Systematic Review, Meta-Analysis, and Omics Exploration
Abstract
Background: Neural precursor cell expressed developmentally downregulated 9 (NEDD9) is considered an important factor in the progression of cancer, acting as a modulator of cellular migration, adhesion, and metastatic potential. Its significance as a prognostic factor, however, remains unclear, which necessitated a comprehensive review and meta-analysis.
Methods: Our study followed the PRISMA guidelines, analyzing studies from major databases including PubMed, Embase, and Cochrane. Our eligibility criteria included studies evaluating NEDD9 expression in relation to cancer prognosis and outcomes such as overall survival (OS), progression-free survival (PFS), disease-free Survival (DFS), recurrence-free survival (RFS), and cancer-specific survival (CSS). We used random-effects and fixed-effect models for meta-analysis, and we validated our findings by comparative analysis using data from external cohorts like The Cancer Genome Atlas (TCGA).
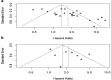
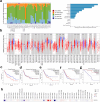
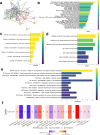
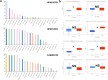
Results: The analysis of 27 studies with 3915 patients demonstrated a significant relationship between NEDD9 expression and poor OS as indicated by the pooled meta-analysis outcome across all included cancers (HR: 1.81, 95% CI: 1.38-2.37). A significant effect on PFS/DFS/RFS/CSS was also found (HR: 2.14, 95% CI: 1.42-3.23). Variations in survival across different types of cancer were indicated by subgroup analysis. NEDD9 expression was correlated with various immune cells across cancer types according to immune infiltration analysis. Protein-protein interaction (PPI) analysis indicated significant interactions involving NEDD9, suggesting mechanisms which influence tumor behavior and response to treatment.
Conclusion: Our results suggest that NEDD9 is a significant prognostic marker in several human cancers. As a result of its central role in cancer progression and prognosis, it presents a promising target for therapeutic interventions. Our study highlights the importance of further research into the biology of NEDD9 and its therapeutic potential.
Keywords: NEDD9 protein; cancer metastasis; gene expression profiling; human; meta-Analysis; oncology; prognostic biomarkers; signal transduction; survival analysis; systematic review; the cancer genome atlas.
Figures




Similar articles
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Prognosis of adults and children following a first unprovoked seizure.Cochrane Database Syst Rev. 2023 Jan 23;1(1):CD013847. doi: 10.1002/14651858.CD013847.pub2. Cochrane Database Syst Rev. 2023. PMID: 36688481 Free PMC article.
-
Optimisation of chemotherapy and radiotherapy for untreated Hodgkin lymphoma patients with respect to second malignant neoplasms, overall and progression-free survival: individual participant data analysis.Cochrane Database Syst Rev. 2017 Sep 13;9(9):CD008814. doi: 10.1002/14651858.CD008814.pub2. Cochrane Database Syst Rev. 2017. PMID: 28901021 Free PMC article.
Cited by
-
Mechanisms of miR-18a-5p Target NEDD9-Mediated Suppression of H5N1 Influenza Virus in Mammalian and Avian Hosts.Vet Sci. 2025 Mar 3;12(3):240. doi: 10.3390/vetsci12030240. Vet Sci. 2025. PMID: 40266966 Free PMC article.
References
-
- Massagué J, Ganesh K. Metastasis-Initiating cells and ecosystems. Cancer Discov. Apr 2021;11(4):971-994. doi:10.1158/2159-8290.Cd-21-0010 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

