Neural network-based predictions of antimicrobial resistance phenotypes in multidrug-resistant Acinetobacter baumannii from whole genome sequencing and gene expression
- PMID: 39540735
- PMCID: PMC11619347
- DOI: 10.1128/aac.01446-24
Neural network-based predictions of antimicrobial resistance phenotypes in multidrug-resistant Acinetobacter baumannii from whole genome sequencing and gene expression
Abstract
Whole genome sequencing (WGS) potentially represents a rapid approach for antimicrobial resistance genotype-to-phenotype prediction. However, the challenge still exists to predict fully minimum inhibitory concentrations (MICs) and antimicrobial susceptibility phenotypes based on WGS data. This study aimed to establish an artificial intelligence-based computational approach in predicting antimicrobial susceptibilities of multidrug-resistant Acinetobacter baumannii from WGS and gene expression data. Antimicrobial susceptibility testing (AST) was performed using the broth microdilution method for 10 antimicrobial agents. In silico multilocus sequence typing (MLST), antimicrobial resistance genes, and phylogeny based on cgSNP and cgMLST strategies were analyzed. High-throughput qPCR was performed to measure the expression level of antimicrobial resistance (AMR) genes. Most isolates exhibited a high level of resistance to most of the tested antimicrobial agents, with the majority belonging to the IC2/CC92 lineage. Phylogenetic analysis revealed undetected transmission events or local outbreaks. The percentage agreements between AMR phenotype and genotype ranged from 70.08% to 89.96%, with the coefficient of agreement (κ) extending from 0.025 and 0.881. The prediction of AST employed by deep neural network models achieved an accuracy of up to 98.64% on the testing data set. Additionally, several linear regression models demonstrated high prediction accuracy, reaching up to 86.15% within an error range of one gradient, indicating a linear relationship between certain gene expressions and the corresponding antimicrobial MICs. In conclusion, neural network-based predictions could be used as a tool for the surveillance of antimicrobial resistance in multidrug-resistant A. baumannii.
Keywords: Acinetobacter baumannii; deep neural network; gene expression; phenotype prediction; whole genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


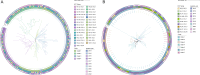


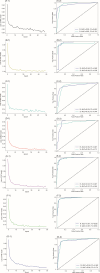

References
-
- Vázquez-López R, Solano-Gálvez SG, Juárez Vignon-Whaley JJ, Abello Vaamonde JA, Padró Alonzo LA, Rivera Reséndiz A, Muleiro Álvarez M, Vega López EN, Franyuti-Kelly G, Álvarez-Hernández DA, Moncaleano Guzmán V, Juárez Bañuelos JE, Marcos Felix J, González Barrios JA, Barrientos Fortes T. 2020. Acinetobacter baumannii resistance: a real challenge for clinicians. Antibiotics (Basel) 9:205. doi:10.3390/antibiotics9040205 - DOI - PMC - PubMed
-
- Mohd Sazlly Lim S, Zainal Abidin A, Liew SM, Roberts JA, Sime FB. 2019. The global prevalence of multidrug-resistance among Acinetobacter baumannii causing hospital-acquired and ventilator-associated pneumonia and its associated mortality: a systematic review and meta-analysis. J Infect 79:593–600. doi:10.1016/j.jinf.2019.09.012 - DOI - PubMed
-
- Amer MM, Mekky HM, Amer AM, Fedawy HS. 2018. Antimicrobial resistance genes in pathogenic Escherichia coli isolated from diseased broiler chickens in Egypt and their relationship with the phenotypic resistance characteristics. Vet World 11:1082–1088. doi:10.14202/vetworld.2018.1082-1088 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 82102436/National Natural Science Foundation of China (NSFC)
- 81401698, 82472335/National Natural Science Foundation of China (NSFC)
- 2020RC066, 2021KY827/Medical and Health Research Project of Zhejiang Province
- 82073610/National Natural Science Foundation of China (NSFC)
- 81871696/National Natural Science Foundation of China (NSFC)
LinkOut - more resources
Full Text Sources
Medical

