The UBA1-STUB1 Axis Mediates Cancer Immune Escape and Resistance to Checkpoint Blockade
- PMID: 39540840
- PMCID: PMC11803397
- DOI: 10.1158/2159-8290.CD-24-0435
The UBA1-STUB1 Axis Mediates Cancer Immune Escape and Resistance to Checkpoint Blockade
Abstract
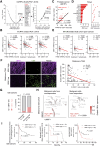
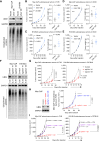
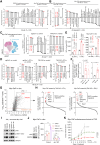
Our study reveals UBA1 as a predictive biomarker for clinical outcomes in ICB cohorts, mediating cancer immune evasion and ICB resistance. We further highlight JAK1 stabilization as a key mechanism of UBA1 inhibition and nominate the UBA1-STUB1 axis as an immuno-oncology therapeutic target to improve the efficacy of ICB.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
W. Zou reports personal fees from CStone, HanchorBio, and NextCure outside the submitted work. A.M. Chinnaiyan reports grants from Prostate Cancer Foundation and NIH during the conduct of the study; personal fees from Medsyn, Ascentage, Esanik Therapeutics, Aurigene Oncology, Rappta Therapeutics, Lynx Dx, Deciphera, and has previously served on the advisory boards of Tempus, EdenRoc, and Flamingo Therapeutics outside the submitted work; and a patent for UBA1 is pending. No disclosures were reported by the other authors.
Figures



References
-
- Janjigian YY, Shitara K, Moehler M, Garrido M, Salman P, Shen L, et al. . First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): a randomised, open-label, phase 3 trial. Lancet 2021;398:27–40. - PMC - PubMed
-
- Sahin U, Oehm P, Derhovanessian E, Jabulowsky RA, Vormehr M, Gold M, et al. . An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature 2020;585:107–12. - PubMed
-
- Sharma P, Pachynski RK, Narayan V, Fléchon A, Gravis G, Galsky MD, et al. . Nivolumab plus ipilimumab for metastatic castration-resistant prostate cancer: preliminary analysis of patients in the CheckMate 650 trial. Cancer Cell 2020;38:489–99.e3. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

