Neutrophil-centric analysis of gastric cancer: prognostic modeling and molecular insights
- PMID: 39540948
- PMCID: PMC11564594
- DOI: 10.1007/s00018-024-05484-w
Neutrophil-centric analysis of gastric cancer: prognostic modeling and molecular insights
Abstract
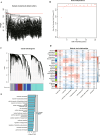
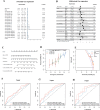
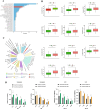
Gastric cancer remains a significant global health concern with poor prognosis. This study investigates the role of neutrophils in gastric cancer progression and their potential as prognostic indicators. Using multi-omics approaches, including Weighted Gene Co-expression Network Analysis (WGCNA), machine learning, and single-cell analysis, we identified neutrophil-associated gene signatures and developed a robust prognostic model. Our findings reveal distinct gastric cancer subtypes based on neutrophil-associated genes, with one subtype showing increased neutrophil infiltration and poorer prognosis. Single-cell analysis uncovered neutrophil-associated alterations in cell composition, gene expression profiles, and intercellular communication within the tumor microenvironment. Additionally, we explored the relationship between neutrophil-associated genes, microbiota composition, and alternative splicing events in gastric cancer. Furthermore, we identified QKI as a key regulator of alternative splicing and demonstrated its role in promoting malignant phenotypes and enhancing TGF-beta signaling and epithelial-mesenchymal transition in gastric cancer cells by wet experiment. Lastly, the role of QKI in the association with drug resistance and the identification of specific agents for treating QKI-associated drug resistance were also explored. This comprehensive study provides novel insights into the complex interplay between neutrophils, the tumor microenvironment, microbiota, alternative splicing and gastric cancer progression, offering potential new targets for therapeutic intervention.
Keywords: Alternative splicing; Drug resistance; Epithelial-mesenchymal transition (EMT); Gastric cancer; Machine learning; Microbiota; Neutrophils; Prognosis model; QKI; Single-cell analysis; TGF-beta signaling; Tumor microenvironment.
© 2024. The Author(s).
Conflict of interest statement
Figures










Similar articles
-
The heterogeneous roles of neutrophils in gastric cancer: scaffold or target?Cell Mol Biol Lett. 2025 Jun 16;30(1):71. doi: 10.1186/s11658-025-00744-4. Cell Mol Biol Lett. 2025. PMID: 40524157 Free PMC article. Review.
-
Integrative analysis of single-cell and bulk RNA sequencing reveals the oncogenic role of ANXA5 in gastric cancer and its association with drug resistance.Front Immunol. 2025 Mar 7;16:1562395. doi: 10.3389/fimmu.2025.1562395. eCollection 2025. Front Immunol. 2025. PMID: 40124374 Free PMC article.
-
Comprehensive Analysis of Alternative Splicing in Gastric Cancer Identifies Epithelial-Mesenchymal Transition Subtypes Associated with Survival.Cancer Res. 2022 Feb 15;82(4):543-555. doi: 10.1158/0008-5472.CAN-21-2117. Cancer Res. 2022. PMID: 34903603 Free PMC article.
-
MicroRNA‑27a‑3p promotes epithelial‑mesenchymal transition by targeting NOVA alternative splicing regulator 1 in gastric cancer.Mol Med Rep. 2020 Mar;21(3):1615-1622. doi: 10.3892/mmr.2020.10949. Epub 2020 Jan 17. Mol Med Rep. 2020. PMID: 32016460
-
RNA-binding protein quaking: a multifunctional regulator in tumour progression.Ann Med. 2025 Dec;57(1):2443046. doi: 10.1080/07853890.2024.2443046. Epub 2024 Dec 23. Ann Med. 2025. PMID: 39711373 Free PMC article. Review.
Cited by
-
Neutrophil-driven tumor inflammation in breast cancer: Pathways and therapeutic implications: A narrative review.Medicine (Baltimore). 2025 Jun 20;104(25):e43024. doi: 10.1097/MD.0000000000043024. Medicine (Baltimore). 2025. PMID: 40550048 Free PMC article. Review.
-
Integrative Analysis of Neutrophil-Associated Genes Reveals Prognostic Significance and Immune Microenvironment Modulation in Cervical Cancer.Biomedicines. 2025 May 30;13(6):1348. doi: 10.3390/biomedicines13061348. Biomedicines. 2025. PMID: 40564066 Free PMC article.
-
The heterogeneous roles of neutrophils in gastric cancer: scaffold or target?Cell Mol Biol Lett. 2025 Jun 16;30(1):71. doi: 10.1186/s11658-025-00744-4. Cell Mol Biol Lett. 2025. PMID: 40524157 Free PMC article. Review.
References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249 - PubMed
-
- Coffelt SB, Wellenstein MD, de Visser KE (2016) Neutrophils in cancer: neutral no more. Nat Rev Cancer 16:431–446 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

