The methyltransferase MLL4 promotes nonalcoholic steatohepatitis by enhancing NF-κB signaling
- PMID: 39542242
- PMCID: PMC11665687
- DOI: 10.1016/j.jbc.2024.107984
The methyltransferase MLL4 promotes nonalcoholic steatohepatitis by enhancing NF-κB signaling
Abstract
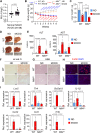
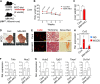
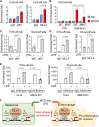
Non-alcoholic fatty liver disease (NAFLD) is a growing health problem worldwide, ranging from non-alcoholic fatty liver (NAFL) to the more severe metabolic non-alcoholic steatohepatitis (NASH). Although many studies have elucidated the pathogenesis of NAFLD, the epigenetic regulatory mechanism from NAFL to NASH remains incompletely understood. The histone H3 lysine 4 methyltransferase, MLL4 (also called KMT2D), is a critical epigenetic transcriptional coactivator that mediates overnutrition-induced steatosis in mice, but its potential role in the progression of NASH remains largely unknown. Here, we show that mice lacking the one allele of the Mll4 gene are resistant to hepatic steatosis, inflammation, and fibrosis in NASH conditions compared to wild-type controls. Transcriptome analysis of the livers of control and Mll4+/- mice identified pro-inflammatory genes regulated by the nuclear factor kappa B (NF-κB) signaling pathway as major target genes of MLL4. We show that MLL4 binds to p65 and that MLL4 is required for NF-κB transactivation. Myeloid-specific Mll4 knockout mice showed an almost complete block of NASH, while hepatocyte-specific Mll4 knockout mice showed mild inhibition of steatosis. Pro-inflammatory M1 polarization is decreased and anti-inflammatory M2 polarization is increased in liver macrophages from myeloid-specific Mll4 knockout mice. Importantly, we show that histone H3-lysine 4 methylation mediated by the MLL4-complex plays a critical role in promoting the expression of Ccl2 in hepatocytes and M1 marker genes in macrophages. Our results demonstrate that MLL4, through the NF-κB-MLL4 regulatory axis, exacerbates steatohepatitis in the context of an inflammatory response and represents a potential therapeutic target for NASH.
Keywords: NF-kappa B (NF-κB); epigenetics; gene expression; hepatocyte; histone methylation; inflammation; macrophage.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interests The authors declare that they have no conflicts of interest with the contents of this article.
Figures




References
-
- Eslam M., Newsome P.N., Sarin S.K., Anstee Q.M., Targher G., Romero-Gomez M., et al. A new definition for metabolic dysfunction-associated fatty liver disease: an international expert consensus statement. J. Hepatol. 2020;73:202–209. - PubMed
-
- Michelotti G.A., Machado M.V., Diehl A.M. NAFLD, NASH and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2013;10:656–665. - PubMed
-
- Day C.P., James O.F. Steatohepatitis: a tale of two "hits"? Gastroenterology. 1998;114:842–845. - PubMed
-
- Kazankov K., Jørgensen S.M.D., Thomsen K.L., Møller H.J., Vilstrup H., George J., et al. The role of macrophages in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Nat. Rev. Gastroenterol. Hepatol. 2019;16:145–159. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

