The circular RNA circNFIX regulates MEF2C expression in muscle satellite cells in spastic cerebral palsy
- PMID: 39542245
- PMCID: PMC11697776
- DOI: 10.1016/j.jbc.2024.107987
The circular RNA circNFIX regulates MEF2C expression in muscle satellite cells in spastic cerebral palsy
Abstract
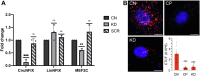
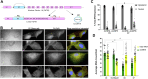
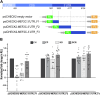
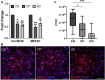
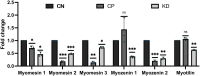
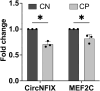
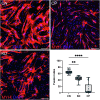
Cerebral palsy (CP) is a pediatric onset disorder with poorly understood molecular causes and progression, making early diagnosis difficult. Circular RNAs are regulatory RNAs that show promise as biomarkers in various diseases but the role of circular RNAs in CP is beginning to be understood. This study identified the role of circNFIX in regulating the expression of myocyte-specific enhancer factor 2C (MEF2C), an important transcription factor for sarcomere development. We found that circNFIX is downregulated in the muscle cells of individuals with CP, and its localization shifts toward the nucleus as visualized using single-molecule resolution imaging. The decreased expression of circNFIX, MEF2C, and MEF2C targets persisted throughout myoblasts to myotubes differentiation, and in the skeletal muscle tissue. Bioinformatic and experimental validation confirmed that circNFIX acts as a sponge for miR373-3p, a microRNA that represses MEF2C translation. In normal muscle, circNFIX derepresses MEF2C translation by sponging miR373-3p, allowing for normal sarcomere generation. In CP, reduced circNFIX expression results in loss of miRNA sponging, leading to lower MEF2C expression and downregulation of sarcomere genes, potentially causing shortened and dysfunctional muscle fibers. Knockdown (KD) of circNFIX reduced myogenic capacity of myoblasts to fuse and form myotubes similar to CP cells evident from the lower fusion index in CP and KD as compared to control myotubes. This is the first study reporting reduction of MEF2C in CP and single-molecule resolution imaging of circNFIX's subcellular distribution and its role in CP, suggesting circNFIX as a potential therapeutic target and biomarker for early CP diagnosis.
Keywords: MEF2C; biomarker; cerebral palsy; circFISH; circNFIX; circular RNA; miR-373-3p; miRNA sponging; regulatory noncoding RNAs; satellite cells; spastic cerebral palsy.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures




References
-
- Lieber R.L., Fridén J. Spasticity causes a fundamental rearrangement of muscle-joint interaction. Muscle Nerve. 2002;25:265–270. - PubMed
-
- Accardo P., Capute A. 3er ed Edition. Paul H. Brookes; Baltimore, MD: 2008. Capute & Accardo’s Neurodevelopmental Disabilities in Infancy and Childhood.
-
- Mockford M., Caulton J.M. The pathophysiological basis of weakness in children with cerebral palsy. Pediatr. Phys. Ther. 2010;22:222–233. - PubMed
-
- Sankar C., Mundkur N. Cerebral palsy-definition, classification, etiology and early diagnosis. Indian J. Pediatr. 2005;72:865–868. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

