Regulation of de novo and maintenance DNA methylation by DNA methyltransferases in postimplantation embryos
- PMID: 39542247
- PMCID: PMC11742614
- DOI: 10.1016/j.jbc.2024.107990
Regulation of de novo and maintenance DNA methylation by DNA methyltransferases in postimplantation embryos
Abstract
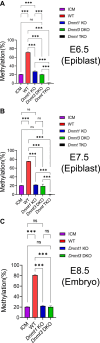
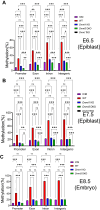
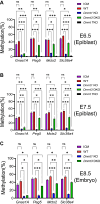
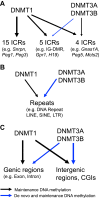
DNA methylation is mainly catalyzed by three DNA methyltransferase (DNMT) proteins in mammals. Usually DNMT1 is considered the primary DNMT for maintenance DNA methylation, whereas DNMT3A and DNMT3B function in de novo DNA methylation. Interestingly, we found DNMT3A and DNMT3B exerted maintenance and de novo DNA methylation in postimplantation mouse embryos. Together with DNMT1, they maintained DNA methylation at some pluripotent genes and lineage marker genes. Germline-derived DNA methylation at the imprinting control regions (ICRs) is stably maintained in embryos. DNMT1 maintained DNA methylation at most ICRs in postimplantation embryos. Surprisingly, DNA methylation was increased at five ICRs after implantation, and two DNMT3 proteins maintained the newly acquired DNA methylation at two of these five ICRs. Intriguingly, DNMT3A and DNMT3B maintained preexisting DNA methylation at four other ICRs, similar to what we found in embryonic stem cells before. These results suggest that DNA methylation is more dynamic than originally thought during embryogenesis including the ICRs of the imprinted regions. DNMT3A and DNMT3B exert both de novo and maintenance DNA methylation functions after implantation. They maintain large portions of newly acquired DNA methylation at variable degrees across the genome in mouse embryos, together with DNMT1. Furthermore, they contribute to maintenance of preexisting DNA methylation at a subset of ICRs as well as in the CpG islands and certain lineage marker gene. These findings may have some implications for the important roles of DNMT proteins in development and human diseases.
Keywords: DNA methylation; DNMT1; DNMT3A; DNMT3B; de novo methylation; genomic imprinting; implantation; imprinting control region (ICR); maintenance methylation; whole-genome bisulfite sequencing (WGBS).
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

Similar articles
-
DNMT1, DNMT3A and DNMT3B proteins are differently expressed in mouse oocytes and early embryos.J Mol Histol. 2017 Dec;48(5-6):417-426. doi: 10.1007/s10735-017-9739-y. Epub 2017 Oct 13. J Mol Histol. 2017. PMID: 29027601
-
Distinct roles of DNMT1-dependent and DNMT1-independent methylation patterns in the genome of mouse embryonic stem cells.Genome Biol. 2015 Jun 2;16(1):115. doi: 10.1186/s13059-015-0685-2. Genome Biol. 2015. PMID: 26032981 Free PMC article.
-
In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b.Mol Cell Biol. 1999 Dec;19(12):8211-8. doi: 10.1128/MCB.19.12.8211. Mol Cell Biol. 1999. PMID: 10567546 Free PMC article.
-
Tissue-specific roles of de novo DNA methyltransferases.Epigenetics Chromatin. 2025 Jan 17;18(1):5. doi: 10.1186/s13072-024-00566-2. Epigenetics Chromatin. 2025. PMID: 39819598 Free PMC article. Review.
-
Using human disease mutations to understand de novo DNA methyltransferase function.Biochem Soc Trans. 2024 Oct 30;52(5):2059-2075. doi: 10.1042/BST20231017. Biochem Soc Trans. 2024. PMID: 39446312 Free PMC article. Review.
Cited by
-
Epigenetic mechanisms involved in hepatocellular carcinoma development and progression.eGastroenterology. 2025 May 4;3(2):e100186. doi: 10.1136/egastro-2025-100186. eCollection 2025. eGastroenterology. 2025. PMID: 40432834 Free PMC article. Review.
-
Unravelling the role of epigenetic regulators during embryonic development of Rhipicephalus microplus.bioRxiv [Preprint]. 2025 Jul 11:2025.07.11.662657. doi: 10.1101/2025.07.11.662657. bioRxiv. 2025. PMID: 40672298 Free PMC article. Preprint.
-
Epigenetic-modification associated hnRNPA3 acts as a prognostic biomarker and promotes malignant progression of HCC.BMC Cancer. 2025 Apr 10;25(1):661. doi: 10.1186/s12885-025-14028-9. BMC Cancer. 2025. PMID: 40211173 Free PMC article.
-
LINE-1 Methylation sustains telomere length in pregnant women: effects on pregnancy failure.Clin Epigenetics. 2025 Jul 23;17(1):130. doi: 10.1186/s13148-025-01937-6. Clin Epigenetics. 2025. PMID: 40702545 Free PMC article.
-
Differentiation of mtDNA Methylation in Tissues of Ridgetail White Prawn, Exopalaemon carinicauda.Animals (Basel). 2025 Jul 11;15(14):2037. doi: 10.3390/ani15142037. Animals (Basel). 2025. PMID: 40723500 Free PMC article.
References
-
- Chen Z., Zhang Y. Role of mammalian DNA methyltransferases in development. Annu. Rev. Biochem. 2020;89:135–158. - PubMed
-
- Dor Y., Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018;392:777–786. - PubMed
-
- Mattei A.L., Bailly N., Meissner A. DNA methylation: a historical perspective. Trends Genet. 2022;38:676–707. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

