Missing microbial eukaryotes and misleading meta-omic conclusions
- PMID: 39543100
- PMCID: PMC11564645
- DOI: 10.1038/s41467-024-52212-w
Missing microbial eukaryotes and misleading meta-omic conclusions
Abstract
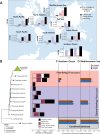
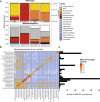
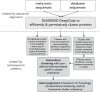
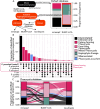
Meta-omics is commonly used for large-scale analyses of microbial eukaryotes, including species or taxonomic group distribution mapping, gene catalog construction, and inference on the functional roles and activities of microbial eukaryotes in situ. Here, we explore the potential pitfalls of common approaches to taxonomic annotation of protistan meta-omic datasets. We re-analyze three environmental datasets at three levels of taxonomic hierarchy in order to illustrate the crucial importance of database completeness and curation in enabling accurate environmental interpretation. We show that taxonomic membership of sequence clusters estimates community composition more accurately than returning exact sequence labels, and overlap between clusters can address database shortcomings. Clustering approaches can be applied to diverse environments while continuing to exploit the wealth of annotation data collated in databases, and selecting and evaluating these databases is a critical part of correctly annotating protistan taxonomy in environmental datasets. We argue that ongoing curation of genetic resources is crucial in accurately annotating protists in in situ meta-omic datasets. Moreover, we propose that precise taxonomic annotation of meta-omic data is a clustering problem rather than a feasible alignment problem.
© 2024. The Author(s).
Conflict of interest statement
Figures

References
-
- Keeling, P. J. & Campo, J. D. Marine protists are not just big bacteria. Curr. Biol.27, R541–R549 (2017). - PubMed
-
- Cuddington, K., Byers, J.E., Wilson, W.G. & Hastings, A. Ecosystem Engineers: Plants to Protists. (Academic Press, 2011).
-
- Caron, D. A., Countway, P. D., Jones, A. C., Kim, D. Y. & Schnetzer, A. Marine protistan diversity. Ann. Rev. Mar. Sci.4, 467–493 (2012). - PubMed
-
- Sherr, E. B. & Sherr, B. F. Significance of predation by protists in aquatic microbial food webs. Antonie Van. Leeuwenhoek81, 293–308 (2002). - PubMed
-
- Worden, A. Z. et al. Environmental science. Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science347, 1257594 (2015). - PubMed

