An interaction network of inner centriole proteins organised by POC1A-POC1B heterodimer crosslinks ensures centriolar integrity
- PMID: 39543170
- PMCID: PMC11564547
- DOI: 10.1038/s41467-024-54247-5
An interaction network of inner centriole proteins organised by POC1A-POC1B heterodimer crosslinks ensures centriolar integrity
Abstract
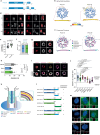
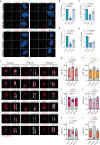
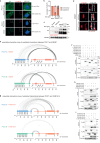
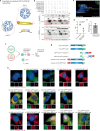
Centriole integrity, vital for cilia formation and chromosome segregation, is crucial for human health. The inner scaffold within the centriole lumen composed of the proteins POC1B, POC5 and FAM161A is key to this integrity. Here, we provide an understanding of the function of inner scaffold proteins. We demonstrate the importance of an interaction network organised by POC1A-POC1B heterodimers within the centriole lumen, where the WD40 domain of POC1B localises close to the centriole wall, while the POC5-interacting WD40 of POC1A resides in the centriole lumen. The POC1A-POC5 interaction and POC5 tetramerization are essential for inner scaffold formation and centriole stability. The microtubule binding proteins FAM161A and MDM1 by binding to POC1A-POC1B, likely positioning the POC5 tetramer near the centriole wall. Disruption of POC1A or POC1B leads to centriole microtubule defects and deletion of both genes causes centriole disintegration. These findings provide insights into organisation and function of the inner scaffold.
© 2024. The Author(s).
Conflict of interest statement
Figures

References
-
- Keller, L. C., Romijn, E. P., Zamora, I., Yates, J. R. & Marshall, W. F. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol.15, 1090–1098 (2005). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

