Enhancing risk stratification models in localized prostate cancer by novel validated tissue biomarkers
- PMID: 39543244
- PMCID: PMC12399428
- DOI: 10.1038/s41391-024-00918-9
Enhancing risk stratification models in localized prostate cancer by novel validated tissue biomarkers
Abstract
Background: Localized prostate cancer (PCa) is a largely heterogeneous disease regarding its clinical behavior. Current risk stratification relies on clinicopathological parameters and distinguishing between indolent and aggressive cases remains challenging. To improve risk stratification, we aimed to identify new prognostic markers for PCa.
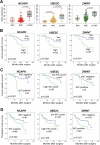
Methods: We performed an in silico analysis on publicly available PCa transcriptome datasets. The top 20 prognostic genes were assessed in PCa tissue samples of our institutional cohort (n = 92) using the NanoString nCounter technology. The three most promising candidates were further assessed by immunohistochemistry (IHC) in an institutional (n = 121) and an independent validation cohort from the EMPACT consortium (n = 199). Cancer-specific survival (CSS) and progression-free survival (PFS) were used as endpoints.
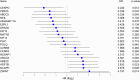
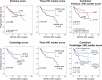
Results: Our in silico analysis identified 113 prognostic genes. The prognostic values of seven of the top 20 genes were confirmed in our institutional radical prostatectomy (RPE) cohort. Low CENPO, P2RX5, ABCC5 as well as high ASF1B, NCAPH, UBE2C, and ZWINT gene expressions were associated with shorter CSS. IHC analysis confirmed the significant associations between NCAPH and UBE2C staining and worse CSS. In the external validation cohort, higher NCAPH and ZWINT protein expressions were associated with shorter PFS. The combination of the newly identified tissue protein markers improved standard risk stratification models, such as D'Amico, CAPRA, and Cambridge prognostic groups.
Conclusions: We identified and validated high tissue levels of NCAPH, UBE2C, and ZWINT as novel prognostic risk factors in clinically localized PCa patients. The use of these markers can improve routinely used risk estimation models.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: BH: Advisory boards for Janssen, Bayer, ABX, Lightpoint, Amgen, MSD, Pfizer, Novartis. Invited speaker for Accord, Astellas, Janssen R&D. Honoraria from Uromed. Research funding from AAA/Novartis, Bristol Myers Squibb, and German Research Foundation. Leadership roles for DKG AUO and DGU. The authors declare no potential conflicts of interest with the present project and publication. Ethics: The study was performed in compliance with the Declaration of Helsinki and the institutional ethics committees approved the study protocol (21-9991-BO and KEK-BE128/2015).
Figures

References
-
- Bray F, Ferlay J, Soerjomataram I. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. A Cancer J Clin. 2018;68:394–424. - PubMed
-
- Chistiakov DA, Myasoedova VA, Grechko AV, Melnichenko AA, Orekhov AN. New biomarkers for diagnosis and prognosis of localized prostate cancer. Semin Cancer Biol. 2018;52:9–16. - PubMed
-
- Jalloh M, Cooperberg MR. Implementation of PSA-based active surveillance in prostate cancer. Biomark Med. 2014;8:747–53. - PubMed
-
- Schröder FH, Hugosson J, Roobol MJ, Tammela TLJ, Ciatto S, Nelen V, et al. ERSPC Investigators. Screening and Prostate-Cancer Mortality in a Randomized European Study. N Engl J Med. 2009;360:1320–8. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

