A comprehensive human embryo reference tool using single-cell RNA-sequencing data
- PMID: 39543283
- PMCID: PMC11725501
- DOI: 10.1038/s41592-024-02493-2
A comprehensive human embryo reference tool using single-cell RNA-sequencing data
Erratum in
-
Author Correction: A comprehensive human embryo reference tool using single-cell RNA-sequencing data.Nat Methods. 2025 Mar;22(3):636. doi: 10.1038/s41592-025-02601-w. Nat Methods. 2025. PMID: 39856235 Free PMC article. No abstract available.
Abstract
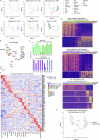
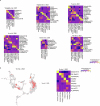
Stem cell-based embryo models offer unprecedented experimental tools for studying early human development. The usefulness of embryo models hinges on their molecular, cellular and structural fidelities to their in vivo counterparts. To authenticate human embryo models, single-cell RNA sequencing has been utilized for unbiased transcriptional profiling. However, an organized and integrated human single-cell RNA-sequencing dataset, serving as a universal reference for benchmarking human embryo models, remains unavailable. Here we developed such a reference through the integration of six published human datasets covering development from the zygote to the gastrula. Lineage annotations are contrasted and validated with available human and nonhuman primate datasets. Using stabilized Uniform Manifold Approximation and Projection, we constructed an early embryogenesis prediction tool, where query datasets can be projected on the reference and annotated with predicted cell identities. Using this reference tool, we examined published human embryo models, highlighting the risk of misannotation when relevant references are not utilized for benchmarking and authentication.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: J. Fu is an editor of NPJ Regenerative Medicine. The other authors declare no competing interests.
Figures














References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

