Diagnosis and prognosis prediction of gastric cancer by high-performance serum lipidome fingerprints
- PMID: 39543322
- PMCID: PMC11628598
- DOI: 10.1038/s44321-024-00169-0
Diagnosis and prognosis prediction of gastric cancer by high-performance serum lipidome fingerprints
Abstract
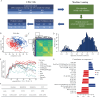
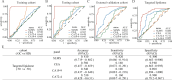
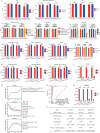
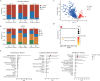
Early detection is warranted to improve prognosis of gastric cancer (GC) but remains challenging. Liquid biopsy combined with machine learning will provide new insights into diagnostic strategies of GC. Lipid metabolism reprogramming plays a crucial role in the initiation and development of tumors. Here, we integrated the lipidomics data of three cohorts (n = 944) to develop the lipid metabolic landscape of GC. We further constructed the serum lipid metabolic signature (SLMS) by machine learning, which showed great performance in distinguishing GC patients from healthy donors. Notably, the SLMS also held high efficacy in the diagnosis of early-stage GC. Besides, by performing unsupervised consensus clustering analysis on the lipid metabolic matrix of patients with GC, we generated the gastric cancer prognostic subtypes (GCPSs) with significantly different overall survival. Furthermore, the lipid metabolic disturbance in GC tissues was demonstrated by multi-omics analysis, which showed partially consistent with that in GC serums. Collectively, this study revealed an innovative strategy of liquid biopsy for the diagnosis of GC on the basis of the serum lipid metabolic fingerprints.
Keywords: Biomarker; Diagnosis; Gastric Cancer; Lipid Metabolism; Prognosis.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests. The authors have applied for patents for the use of the serum lipid metabolic signature to diagnose and predict biosamples.
Figures






References
-
- Chen H, Zhou H, Liang Y, Huang Z, Yang S, Wang X, She Z, Wei Z, Zhang Q (2023a) UHPLC-HRMS-based serum untargeted lipidomics: phosphatidylcholines and sphingomyelins are the main disturbed lipid markers to distinguish colorectal advanced adenoma from cancer. J Pharm Biomed Anal 234:115582 - DOI - PubMed
MeSH terms
Substances
Grants and funding
- 2022yfa1105300/MOST | National Key Research and Development Program of China (NKPs)
- 82341010; 82273241; 8197625; 82103643; 32201217;/MOST | National Natural Science Foundation of China (NSFC)
- 2023B1515040030/Guangdong Basic and Applied Basic Research Foundation
- XLYC2002035/Liaoning Revitalization Talents Program ()
- SZSM202211017/| Sanming Project of Medicine in Shenzen Municipality (Sanming Project of Medicine in Shenzhen)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

