Identification of novel PAD2 inhibitors using pharmacophore-based virtual screening, molecular docking, and MD simulation studies
- PMID: 39543332
- PMCID: PMC11564549
- DOI: 10.1038/s41598-024-78330-5
Identification of novel PAD2 inhibitors using pharmacophore-based virtual screening, molecular docking, and MD simulation studies
Abstract
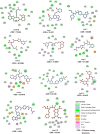
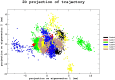
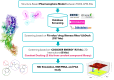
In the realm of epigenetic regulation, Protein arginine deiminase 2 (PAD2) stands out as a therapeutic target due to its significant role in neurological disorders, rheumatoid arthritis (RA), multiple sclerosis (MS), and various cancers. To date, no in silico studies have focused on PAD2 for lead compound identification. Therefore, we conducted structure-based pharmacophore modeling, virtual screening, molecular docking, molecular dynamics (MD) simulations, and essential dynamics studies using PCA and free energy landscape analyses to identify repurposed drugs and selective inhibitors against PAD2. The best pharmacophore model, 'Pharm_01,' had a selectivity score of 10.485 and an excellent ROC curve quality of 0.972. Pharm1 consisted of three hydrogen bond donors (HBD) and two hydrophobic (Hy) features (DDDHH). A virtual screening of about 9.2 million compounds yielded 2575 hits using a fit value threshold of 2.5 and drug-likeness criteria. Molecular docking identified the top ten molecules, which were verified using MD simulations. Stability was verified using MM-PBSA studies, whereas conformational differences were investigated using PCA and free energy landscape analyses. Two hits (Leads 1 and 2) from the DrugBank dataset showed promise for repurposing as PAD2 inhibitors, while one hit compound (Lead 8) from the ZINC database emerged as a novel PAD2 inhibitor. These findings indicate that the discovered compounds may be potent PAD2 inhibitors, necessitating additional preclinical and clinical research to produce viable treatments for cancer and neurological disorders.
Keywords: Drug repurposing; MD simulation; PAD2; PCA analyses; Structure-based Pharmacophore Model; Virtual screening.
© 2024. The Author(s).
Conflict of interest statement
Figures







References
-
- Rogers, G. E., Harding, H. W. J. & Llewellyn-Smith, I. J. The origin of citrulline-containing proteins in the hair follicle and the chemical nature of trichohyalin, an intracellular precursor. Biochim. Biophys. Acta BBA Protein Struct.495, 159–175 (1977). - PubMed
-
- Rogers, G. E. & Simmonds, D. H. Content of citrulline and other amino-acids in a protein of hair follicles. Nature182, 186–187 (1958). - PubMed
-
- Mansouri, P. et al. Peptidylarginine deiminase (PAD): A promising target for chronic diseases treatment. Int. J. Biol. Macromol.278, 134576 (2024). - PubMed
-
- Yuzhalin, A. E. Citrullination in cancer. Cancer Res.79, 1274–1284 (2019). - PubMed
MeSH terms
Substances
Grants and funding
- DST/INSPIRE/03/2016/000026/Department of Science and Technology, Ministry of Science and Technology, India
- DBT/2021-22/CBR/1593/Department of Biotechnology, Ministry of Science and Technology, India
- BT/PR40153/BTIS/137/8/2021/Department of Biotechnology, Ministry of Science and Technology, India
- IoE/2023-24/12/FRP/Dr. B. R. Ambedkar Center for Biomedical Research and the Institute of Eminence, University of Delhi
LinkOut - more resources
Full Text Sources

