Molecular profiling reveals novel therapeutic targets and clonal evolution in ovarian clear cell carcinoma
- PMID: 39543535
- PMCID: PMC11566382
- DOI: 10.1186/s12885-024-13125-5
Molecular profiling reveals novel therapeutic targets and clonal evolution in ovarian clear cell carcinoma
Abstract
Background: Ovarian clear cell carcinoma (OCCC) has a disproportionately high incidence among women in East Asia. Patients diagnosed with OCCC tend to experience worse clinical outcomes than those with high-grade serous carcinoma (HGSC) at advanced stages. The unfavorable prognosis of OCCC can be partly attributed to its frequent resistance to conventional chemotherapy. Within a precision medicine framework, we sought to provide a comprehensive molecular characterization of OCCC using whole-exome sequencing to uncover potential molecular targets that may inform novel therapeutic strategies.
Methods: We performed whole-exome sequencing analysis on tumor-normal paired samples from 102 OCCC patients. This comprehensive genomic characterization of a substantial cohort of OCCC specimens was coupled with an analysis of clonal progression.
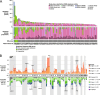
Results: On analyzing 102 OCCC samples, ARID1A (67%) and PIK3CA (49%) emerged as the most frequently mutated driver genes. We identified tier 1 or 2 clinically actionable molecular targets in 40% of cases. This included DNA mismatch repair deficiency (n = 1), as well as BRCA2 (n = 1), PIK3CA (n = 36), KRASG12C (n = 1), and ATM (n = 4) mutations. Furthermore, 45% of OCCC samples displayed ARID1A biallelic loss. Interestingly, we identified previously unreported mutations in the 5' untranslated region of the TERT gene that harbored an adverse prognostic significance. Clock-like mutational processes and activated APOBECs were major drivers of somatic point mutations. Mutations arising from DNA mismatch repair deficiency were uncommon. Reconstruction of clonal evolution revealed that early genetic events likely driving tumorigenesis included mutations in the ARID1A, PIK3CA, TERT, KRAS, and TP53 genes.
Conclusions: Our study provides a comprehensive characterization of the genomic landscape and clonal evolution in OCCC within a substantial cohort. These findings unveil potentially actionable molecular alterations that could be leveraged to develop targeted therapies.
Keywords: TERT mutation; Clinical actionability; Clonal evolution; Mutational signature; Ovarian clear cell carcinoma; Whole-exome sequencing.
© 2024. The Author(s).
Conflict of interest statement
Figures






References
-
- Cabasag CJ, Fagan PJ, Ferlay J, Vignat J, Laversanne M, Liu L, van der Aa MA, Bray F, Soerjomataram I. Ovarian cancer today and tomorrow: A global assessment by world region and Human Development Index using GLOBOCAN 2020. Int J Cancer. 2022;151(9):1535–41. - PubMed
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–49. - PubMed
-
- Ku FC, Wu RC, Yang LY, Tang YH, Chang WY, Yang JE, Wang CC, Jung SM, Lin CT, Chang TC, et al. Clear cell carcinomas of the ovary have poorer outcomes compared with serous carcinomas: Results from a single-center Taiwanese study. J Formos Med Assoc. 2018;117(2):117–25. - PubMed
-
- Chan JK, Teoh D, Hu JM, Shin JY, Osann K, Kapp DS. Do clear cell ovarian carcinomas have poorer prognosis compared to other epithelial cell types? A study of 1411 clear cell ovarian cancers. Gynecol Oncol. 2008;109(3):370–6. - PubMed
-
- del Carmen MG, Birrer M, Schorge JO. Clear cell carcinoma of the ovary: a review of the literature. Gynecol Oncol. 2012;126(3):481–90. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

