Investigation of multiple sclerosis-related pathways through the integration of genomic and proteomic data
- PMID: 39544199
- PMCID: PMC11563213
- DOI: 10.7717/peerj.11922
Investigation of multiple sclerosis-related pathways through the integration of genomic and proteomic data
Abstract
Background: Multiple sclerosis (MS) has a complex pathophysiology, variable clinical presentation, and unpredictable prognosis; understanding the underlying mechanisms requires combinatorial approaches that warrant the integration of diverse molecular omics data.
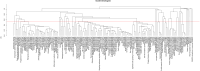
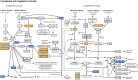
Methods: Here, we combined genomic and proteomic data of the same individuals among a Turkish MS patient group to search for biologically important networks. We previously identified differentially-expressed proteins by cerebrospinal fluid proteome analysis of 179 MS patients and 42 non-MS controls. Among this study group, 11 unrelated MS patients and 60 independent, healthy controls were subjected to whole-genome SNP genotyping, and genome-wide associations were assessed. Pathway enrichment analyses of MS-associated SNPs and differentially-expressed proteins were conducted using the functional enrichment tool, PANOGA.
Results: Nine shared pathways were detected between the genomic and proteomic datasets after merging and clustering the enriched pathways. Complement and coagulation cascade was the most significantly associated pathway (hsa04610, P = 6.96 × 10-30). Other pathways involved in neurological or immunological mechanisms included adherens junctions (hsa04520, P = 6.64 × 10-25), pathogenic Escherichia coli infection (hsa05130, P = 9.03 × 10-14), prion diseases (hsa05020, P = 5.13 × 10-13).
Conclusion: We conclude that integrating multiple datasets of the same patients helps reducing false negative and positive results of genome-wide SNP associations and highlights the most prominent cellular players among the complex pathophysiological mechanisms.
Keywords: Bioinformatics; Genomics; Multiple sclerosis; Proteomics.
©2021 Everest et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures
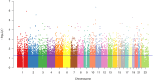
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

